Abstract
The Protein Movie Generator (PMG) is an online service able to generate high-quality pictures and animations for which one can then define simple storyboards. The PMG can therefore efficiently illustrate concepts such as molecular motion or formation/dissociation of complexes. Emphasis is put on the simplicity of animation generation. Rendering is achieved using Dino coupled to POV-Ray. In order to produce highly informative images, the PMG includes capabilities of using different molecular representations at the same time to highlight particular molecular features. Moreover, sophisticated rendering concepts including scene definition, as well as modeling light and materials are available. The PMG accepts Protein Data Bank (PDB) files as input, which may include series of models or molecular dynamics trajectories and produces images or movies under various formats. PMG can be accessed at http://bioserv.rpbs.jussieu.fr/PMG.html.
INTRODUCTION
Most of today's biological and biochemical studies emphasize molecular function; this often implies considering molecular motions or interactions. Visualization of such processes has long been acknowledged as of major importance. Already in the mid 1960s, Cyrus Levinthal and his co-workers produced the first molecular movie (see http://www.umass.edu/molvis/francoeur/movgallery/moviegallery.html) using their ‘model-building’ program (1). Since then, a huge effort has been put into developing computer graphics systems and software to investigate macromolecular structure and function. As imaging has steadily become a major requirement for scientific communication and teaching, the field continues to grow and new software is regularly developed, e.g. SwissPDBViewer (2), VMD (3) PyMol (http://pymol.sourceforge.net), Yasara (http://www.yasara.org), Jmol (http://jmol.sourceforge.net) and many others.
While quality of software and images produced have been increasing steadily, the complexity of concepts and of software can make the task of producing these high-quality pictures or movies difficult to the inexperienced user. To overcome this difficulty, several online services such as Aismig (4), Indie Molecular Movies (http://molbio.info.nih.gov/structbio/indie.html), Molray (5), MovieMaker (6), pdb2mgif (7), POLYVIEW-3D (8) or Protein Picture Generator (PPG) (9) have been developed. They are based on various molecular graphics programs, such as PyMol, RasMol (10), or Dino (http://www.dino3d.org), that are coupled to free 3D rendering engines such as Molscript (11), Raster3D (12) or POV-Ray (http://www.povray.org) to improve the quality of the pictures in terms of photorealism.
It appears that one sticking point for online imaging services is the design of an interface to supervise image production. Ultimately, it would be desirable for such services to generate complex molecular pictures and animations, while controlling numerous parameters at different stages of image production (molecular representation, view control, rendering control, scene control including light, camera, background, etc.). The user should then be able to define a storyboard for animation, including cinematic control. However, in order to propose a comprehensive way for the inexperienced user to generate images, choices and hierarchies must be defined amongst the various possible parameters and selections. The diversity and quality of the molecular representations produced by the software strongly depend on these design choices. So far, Aismig, Molray, pdb2mgif or PPG can produce images, which display several types of representations using various color codes to highlight particular molecular features. In contrast, MovieMaker is more oriented toward the production of animations and can produce short movies illustrating a wide range of protein motions or other dynamic processes. However, less molecular representations can be displayed at one time. In addition, MovieMaker offers little control over the animation. Indeed, none of these services offer complex animation control. For instance, there is no control over the illustration of molecular recognition processes involving several partners.
In order to address the need to design animation storyboards and to take into account the requirements of multilevel molecular representations, we have developed the Protein Movie Generator (PMG). The PMG service relies on concepts previously introduced by the PPG and significantly extends them toward macromolecular movie generation. Namely, it proposes movie generation including several moving molecules for which it is possible to define a scenario considering move order, initial and final positions and/or orientations, as well as several useful camera moves. Since a ray-tracing technique was included in the rendering, PMG can generate images with a good level of photorealism. Nevertheless, the design approach of the PMG form aims to avoid technical considerations for simplicity's sake.
The PMG service can therefore address quite simply a large number of requirements in molecular imaging, ranging from static pictures to simple animations (namely rotations or rock), and complex animation such as ‘docking’ movies, model slideshows and molecular motions or dynamics, while combining various representations and coloring schemes.
DESIGN OF PMG
The PMG interface is designed with respect to experience acquired from the PPG, and from discussions with users. Some of the wishes expressed by users can prove contradictory, e.g. the ability to produce a movie in a few clicks, and the need to access to as many parameters as possible. A point often mentioned involved the possibility of having some interactive control over the orientation of the molecule. Finally, since movie generation can be time consuming, it seemed desirable to have some means to produce movies of macromolecular prototype easily and quickly.
Technical choices
As a consequence, technical choices related to the rendering software were driven by several considerations:
the need of scriptable rendering,
the ability to use different types of molecular representations and coloring schemes at the same time, in order to obtain a high level of scientific information in the images,
the ability to produce high-quality image rendering. We therefore considered the possibility of using ray-tracing.
The Dino software, which is the core of PPG, can define and manage independent molecular objects using various molecular representations. Since Dino is scriptable and can be interfaced with POV-ray, we have chosen to keep using it in PMG. Dino batch rendering is possible using the X virtual frame buffer (Xvfb).
The overall PMG processing is as follows: the cgi form generates a first level script for Dino, which in turn generates a second level script for POV-ray. In fact, the PMG uses Megapov (http://megapov.inetart.net), a clone of POV-ray, which offers interesting features (see below). PMG allows production of renderings using Dino or POV-ray as stand-alone units. Moreover, some ancillary programs are required for some representations, e.g. computation of molecular surfaces [msms (13)] or of secondary structures [stride (14)]. A challenge in setting up an image generation web server is related to the specification of 3D orientation of molecules by the user. For this purpose, we have elected Jmol as an ancillary tool since its applet version provides interactive 3D visualization. Finally, the image post-processing involves the ‘convert’ program (part of the ImageMagick package http://www.imagemagick.org/script/index.php) for title and subtitles setup, and conversion to requested image formats, including animated gif. However, to produce avi formatted movies, we chose to use the mencoder software, included in the MPlayer package at http://www.mplayerhq.hu/design7/news.html).
User interface
The PMG interface is organized as two services. The main service provides access to the various parameters controlling molecular representation, scene, scenarios, etc. The ancillary service, based on Jmol, allows interactive preselection of molecule orientation. Selected orientations can then be exported to the main form.
The main interface of the PMG is organized in different sections, addressing various aspects of the movie production task. The top section covers data input. The next section, i.e. the main control section, is divided in: (i) the default representation for the complete structure (sphere, cartoon, lines, molecular surface and coloring patterns) for which parameters can be set independently, (ii) the scene section which allows the user to center and focus the scene on some part of the structure, and to include several ready-made backgrounds (see http://bioserv.rpbs.jussieu.fr/~autin/help/bg.html, based on http://www.f-lohmueller.de/pov_tut/addon/insert0.htm), (iii) the animation section which proposes pre-defined scenarios and camera behavior (static or dynamic) as well as a limited number of animation controls (number of frames, step and delay). Further sections control numerous additional secondary parameters, such as specific selection of atoms, residues and chains by the user.
Special points on the PMG concern animation. Several types of animations, using the PDB (15) format, are available, including: four basic types of animation (Rock, Xrot, Yrot and Ztrans), visualization of molecular dynamics (which is accessible by uploading a trajectory file), slide shows of models (for files containing more than one model, such as NMR structures) and ‘docking’ movies. This latter type of animation covers both docking of protein chains, and ligand docking.
The program offers many more possibilities: model animation can be used for NMR data, but also for visualization of docking results as produced for instance by the ClusPro (16) server, molecular dynamics as accessible in the Molecular Dynamics Extended Library (http://mmb.pcb.ub.es/MODEL), molecular motions produced from normal mode analysis [see (17,18) for some examples] or from morphing two structures as stored in the Database of Macromolecular Movements (19). For files containing multiple chains, animation is possible for any kind of binding or unbinding process. Using the PQS database (20) or the PITA server (21), both of which compute quaternary structures according the crystallographic symmetry, in combination with PMG, it is therefore possible to illustrate the oligomerization of many proteins stored in the PDB. Finally, to illustrate protein-ligand interactions, the PMG also relies on the definition of a ligand using the HETATM fields of the PDB file format. Further details about these modes are available in the corresponding online help pages of the PMG (for details see http://bioserv.rpbs.jussieu.fr/PMG.html).
All these animation possibilities are supplemented by several camera animation scenarios. Six types of camera use have been defined, and imply automatized focus adaptation. Three of these are static (simple, wide and ultra-wide) and three are dynamic (Zoom, rotY and rotZ). This further increases possibilities for animation.
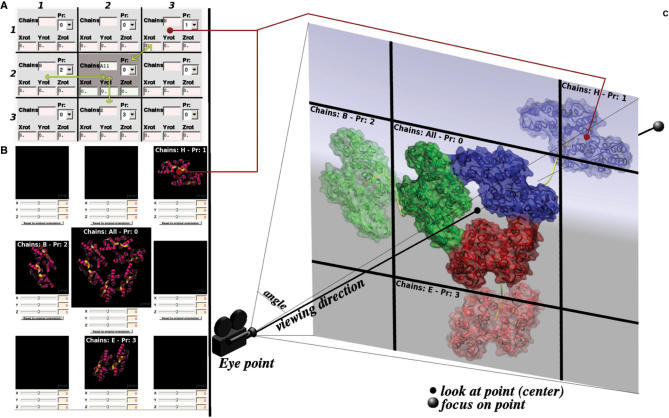
A topic of particular importance is the definition of storyboards. As illustrated in Figure 1A, we chose to virtually split the screen space in a 3 × 3 matrix. For each of the cells in this matrix, it is possible to assign particular chain(s), or the ligand, and to set for each of these a particular molecular orientation and a rank defining the running order in an event sequence. Hence, it is possible to have control over nine groups of objects (multiple selection of chains), eight of which depict a dynamic state in the animation, and the last one corresponding either to the starting state or the final state of the animation (defined in the 3 × 3 matrix by the ‘All’ keyword, see Figure 1A). Once objects are attributed to each cell, it is possible to use the Jmol preview function in order to assign preferred orientation to each group of objects according to their layout in the 3 × 3 matrix (see Figure 1A and B). For example, Figure 1C depicts the oligomerization process defined in Figure 1A and B. Each object is translated and reoriented according to values in the 3 × 3 matrix, and then undergoes a movement toward the final state, i.e. in this case the bound oligomer. To make the motion more realistic, object motion is guided by a spline, which is randomly generated around a straight trajectory (yellow curves in Figure 1C). The scene's center, focus and angle (which depend on the camera option) are automatically set by PMG, but can be defined by the user (see Figure 1C). It is also possible to specify a number of frames, and a step. The meaning of these parameters varies according to the type of animation selected (see the help pages).
Figure 1.
PMG storyboard definition panel. The screen is divided into a virtual matrix of 3 × 3 cells to position objects. It is possible to assign chains or ligand to each cell and to specify their behavior. (A) Assignments are propagated to the orientation previewer (B) and the rendering engine (C). This example shows how to use the panel (A) in order to produce a trimerization. The final state (defined by the ‘All’ key word) is set in the center of the scene (middle cell 2.2.). The starting position of each chain is set by assigning each of them to a different cell (H in 1.3., B in 2.1 and E in 3.2). Once the chains are assigned to cells, it is possible to specify the starting and final orientations by assigning rotations values for each cell (Xrot, Yrot and Zrot fields). To help, it is possible to launch the previewer (B). The timeline of the movements is defined by assigning a rank—or Priority—to each cell set (Pr fields). Here, chain H will move first, then chain B, and finally chain E. These moves are sketched using yellow arrows in (A) and yellow splines in (C). Elementary move magnitude is automatically defined by PMG. Finally, it is possible to change the angle, center and focus of the camera (C), which leads to possibly focusing on a particular region of the macromolecule.
Figure 2.
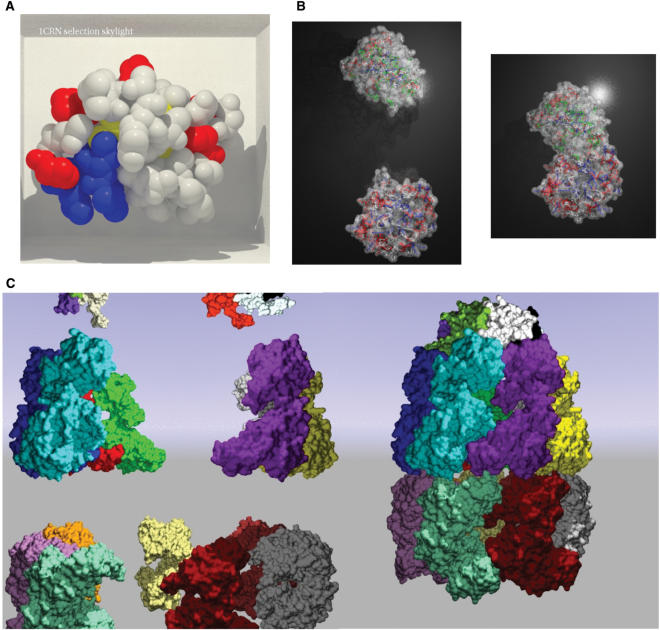
Example of PMG output. (A) Photorealism: Crambin (PDB code 1CRN) displayed in a box using atom spheres. The rendering is done using the ‘Skylight’ option. (B) Simultaneous multiple representations: two chains isomerase (PDB code 1TIM). The chain A (bottom) is displayed using a backbone spline colored according to the secondary structure, and using a 50% transparent gray surface. A selection of a subset of amino-acids (ASP, GLU, ASN, GLN, LYS and HIS) is drawn using a different representation (ball and sticks colored according to atom charge). The chain B is displayed using a backbone trace colored according to the secondary structure and using a 50% transparent gray surface. A second user selection over different amino-acids (ALA, CYS, PHE, GLY, ISO, LEU and MET) is displayed as ball and sticks and colored according to hydrophobicity. The background is a z-plane textured with material reflection. (C) Large protein assembly animation: This example depicts the oligomerization of the chaperonin Groel–Groes complex, PDB code 1AON (starting position = left image, final position = right image). PMG can handle this large protein assembly (more than 8.000 residues for 21 chains), first by using an ultra wide-angle camera (‘ultra-wide’ option), and then by grouping chains as objects, i.e. by assigning several chains to the same cell, for instance in cell 3.3 Chains: K, L, M and in cell 2.3 Chains: D, E, F. The background is a blue to green sky sphere (‘spheresky’). The complete animation is available at the PMG gallery (http://bioserv.rpbs.jussieu.fr/~autin/help/gallery.html).
Finally, the use of Megapov (a clone of POV-ray) for movie generation allows sophisticated control over the animation clock (see http://www.geocities.com/SiliconValley/Lakes/1434/clockmod.html), which in turn allows non-linear kinematics (the animation starts slowly, speeds up and then slows down). The corresponding parameters are hidden to the user.
To summarize the interface, over the ∼200 parameters set in PMG, only half of these are accessible to the user. Furthermore, using only the 30 main parameters (covering default representation, scene and animation parameters), the user can easily produce images or animations of high quality.
INPUT/OUTPUT
Data input can be on the form of typical PDB files (single chains, multichains and multimodels), but also on the form of molecular dynamics trajectories in several formats including Gromacs (.xtc), Charmm (.dcd) and CNS (.crd). The PMG can produce static images (PNG, JPG or PS format) as well as animations (GIF and AVI format).
PERFORMANCES
Most basic rendering requests require no more than 10 min of computational time. However, computations including transparency of surfaces can require several hours. Finally, one of the most time consuming parameters relates to the material reflection. If this is to be considered, computational cost can increase up to days or weeks. In order not to overload the service, depending on parameters and size of the input, it is possible that PMG will refuse to process some requests. It is however possible to contact the authors for such particular requests.
The Figure 2 presents two examples of static image production, and one example of oligomerization (only the starting and the final state). For further animation examples see the PMG gallery at http://bioserv.rpbs.jussieu.fr/~autin/help/gallery.html, which provides examples of requests as well as information about cpu-time.
DISCUSSION AND FUTURE WORK
The PMG, via a large number of parameters, can serve the non-specialist as well as the specialist. It covers a large variety of rendering possibilities to illustrate various molecular topics. One of its main feature is undoubtedly its strong animation capabilities.
Nevertheless, several limitations appeared during the design of the PMG. The first, mentioned above, concerns some rendering parameters, which are highly demanding in cpu-time, in particular parameters related to the calculation of light reflection. For this reason, the PMG does not allow using all the photorealistic capabilities of the POV-ray engine—although we tried to overcome this limitation using special light functions (such as the ‘SkySphere’ light option, see the gallery). Among the possibilities that have not been explored are object material modeling, and concepts such as ambient light occlusion which allow even more realistic molecular rendering (see for instance http://qutemol.sourceforge.net), which really requires huge amounts of computational time. Such limitations could be partly addressed since POV-ray has intrinsic capabilities for parallel computation. Another limitation comes from the propagation of the default representation. It would be desirable to allow independent rendering schemes for objects, thus escaping the default representation. This is partly possible using the user selection facilities, but still leaves room for improvement. Finally, more complex camera and object moves could be defined, and some possibilities could be given for scene background, such as the use of user images or textures. Overall, however, the PMG's present choices will make it easy to use for many scientific or educational applications.
ACKNOWLEDGEMENTS
The authors wish to thank the many users that have contributed to guide the development of the PMG. The authors thank J. Becq and J. Nelson for their help regarding linguistic issues. Funding to pay the Open Access publication charges for this article was provided by INSERM and University Paris Diderot-Paris 7.
Conflict of interest statement. None declared.
REFERENCES
- 1.Levinthal C. Molecular Model-Building by Computer. Sci. Am. 1966;214:42–52. doi: 10.1038/scientificamerican0666-42. [DOI] [PubMed] [Google Scholar]
- 2.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 3.Humphrey W, Dalke A, Schulten K. VMD—Visual Molecular Dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 4.Bohne-Lang A, Groch WD, Ranzinger R. AISMIG—an interactive server-side molecule image generator. Nucleic Acids Res. 2005;33:W705–W709. doi: 10.1093/nar/gki438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mark H, Alwyn JT. Molray—a web interface between O and the POV-Ray ray tracer. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1201–1203. doi: 10.1107/s0907444901007697. [DOI] [PubMed] [Google Scholar]
- 6.Maiti R, Van Domselaar GH, Wishart DS. MovieMaker: a web server for rapid rendering of protein motions and interactions. Nucleic Acids Res. 2005;33:W358–W3562. doi: 10.1093/nar/gki485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohne A. PDB2multiGIF: a web tool to create animated images of molecules. J. Mol. Model. 1998;4:344–346. [Google Scholar]
- 8.Porollo A, Adamczak R, Meller J. POLYVIEW: a flexible visualization tool for structural and functional annotations of proteins. Bioinformatics. 2004;20:2460–2462. doi: 10.1093/bioinformatics/bth248. [DOI] [PubMed] [Google Scholar]
- 9.Binisti C, Salim AA, Tuffery P. PPG: online generation of protein pictures and animations. Nucleic Acids Res. 2005;33:W320–W323. doi: 10.1093/nar/gki392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 11.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 12.Merritt EA. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 13.Sanner MF, Olson AJ, Spehner J-C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 14.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 15.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 17.Suhre K, Sanejouand YH. ElNemo: a normal mode web-server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flores S, Echols N, Milburn D, Hespenheide B, Keating K, Lu J, Wells S, Yu EZ, Thorpe M, et al. The database of macromolecular motions: new features added at the decade mark. Nucleic Acids Res. 2006;34:D296–D301. doi: 10.1093/nar/gkj046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 21.Ponstingl H, Kabir T, Thornton JM. Automatic inference of protein quaternary structure from crystals. J. Appl. Cryst. 2003;36:1116–1122. [Google Scholar]