Abstract
The web application MAGMA provides a simple and intuitive interface to identify differentially expressed genes from two-channel microarray data. While the underlying algorithms are not superior to those of similar web applications, MAGMA is particularly user friendly and can be used without prior training. The user interface guides the novice user through the most typical microarray analysis workflow consisting of data upload, annotation, normalization and statistical analysis. It automatically generates R-scripts that document MAGMA's entire data processing steps, thereby allowing the user to regenerate all results in his local R installation. The implementation of MAGMA follows the model-view-controller design pattern that strictly separates the R-based statistical data processing, the web-representation and the application logic. This modular design makes the application flexible and easily extendible by experts in one of the fields: statistical microarray analysis, web design or software development. State-of-the-art Java Server Faces technology was used to generate the web interface and to perform user input processing. MAGMA's object-oriented modular framework makes it easily extendible and applicable to other fields and demonstrates that modern Java technology is also suitable for rather small and concise academic projects. MAGMA is freely available at www.magma-fgcz.uzh.ch.
INTRODUCTION
Microarrays have become a standard research tool for gene expression analysis and a number of established solutions exist for the subsequent data analysis. The R language and the corresponding Bioconductor packages (1) have established themselves as the reference platform for the publication and implementation of the latest bioinformatic algorithms. The available packages provide a comprehensive functionality for all aspects of microarray data analysis. Data preprocessing, normalization, explorative data mining, differential expression analysis, pathway and GO analysis can be done as well as even more sophisticated analyses that elucidate mechanisms of transcriptional regulation, identify gene networks or perform text mining. However, using the R programming language requires computing skills that go beyond the skills of a typical academic life science researcher and microarray expert. As an alternative, a range of web applications exist that allow users to perform microarray data analysis with their web browser. The Bioinformatics Links Directory (http://bioinformatics.ubc.ca/resources/links_directory) gives an overview of the available web servers. Intermediate to experienced users can exploit these applications for the analysis of their data. However, for a biologist or medical researcher with no or little experience in microarray analysis, the majority of these tools are demanding and require too much prior knowledge about file formats, preprocessing options and data characteristics to be used right away without any further assistance.
The web application MAGMA fills this gap by providing a quickstart to two-channel microarray data analysis especially to novice microarray users. It requires only the essential minimum of user input that is needed to be able to run the most frequently used microarray analysis task: the identification of differentially expressed genes. The web interface is specifically designed to be easy and intuitive to use without requiring users to have previous training. Nevertheless, MAGMA performs all the necessary steps to correctly process the data and compute the differentially expressed genes from two-channel microarray data. It is intended to be run directly after the hybridization and image quantification and therefore satisfies the need of many researchers to obtain an instant overview of the amount of differential expression and to get an initial list of regulated genes.
MAGMA's architecture follows the Model-View-Controller design pattern (http://en.wikipedia.org/wiki/Model-view-controller). With this design we achieve a clear separation of the user interface (web pages) from the application logic (Servlets), and the statistical data processing that is delegated to R and Bioconductor. The data processing relies on a framework that models an entire microarray data analysis as a series of atomic processing steps. The results of the individual steps are persistent and offer thus a history that allows users to go back to any previous processing step. Given the modular design of the architecture and the data processing, MAGMA's functionality can be extended by simply implementing additional processing steps without the need to change the code of the core engine or the existing processing steps. The generality of the data processing model also makes the framework suitable for other applications requiring completely different data processing steps.
Examples of other web applications with similar or more extensive microarray analysis functionalities include CARMAweb (2), ArrayQuest (3), Expression Profiler (4), ExpressYourself (5), RACE (6), ArrayPipe (7), GenePublisher (8), SNOMAD (9), GEPAS (10), WebArray (11) and MIDAW (12). Of these applications, only CARMAweb, RACE and GenePublisher generate R-Scripts which allow the user to reproduce the processing on his computer locally. Furthermore, only CARMAweb relies also on a modern Java architecture, however it does not yet make use of the Java Server Faces (JSF) technology which takes from the developers the burden to implement standard GUI functionality. Generally, these web applications aim at providing a comprehensive set of microarray data analysis functionalities. However, to achieve this purpose they compromise on the ease-of-use for first-time users who, instead of a full blown analysis, simply want to get a fast overview over their list of regulated genes. MAGMA on the other hand focuses specifically on this functionality and lets users compute the list of differentially expressed genes without detouring. We assume that users will perform a subsequent in-depth analysis with a dedicated software package (e.g. the academic TM4 suite: http://www.tm4.org/) or a commercial product such as GeneSpring (Agilent), Resolver (Rosetta), Expressionist (Genedata) or similar. MAGMA does, therefore, not intend to replace these applications but complements them by allowing users to carry out a quick initial analysis without the need of correctly filling in detailed LIMS information or setting up gene annotation. In our experience, first-time users are overwhelmed by the huge number of analyses and options these applications offer, and do not get to any result within a reasonable time.
METHODS
All data analysis operations in MAGMA are performed using the R language and Bioconductor packages. By relying on Bioconductor's limma package (13), MAGMA accurately models and analyzes the typical two-group comparisons in reference and dye-swap experimental designs. In some cases, Bioconductor functions have been extended with additional error checks because the available functions would not test appropriately for the validity of the input and the correctness of the result. All data operations are tracked in an R-script, making them accessible to the user for documentation or reuse. Users familiar with R may take the generated R-script, paste it into their local R installation and reproduce the entire analysis on their local computer. This could also serve as a starting point for a subsequent more advanced analysis.
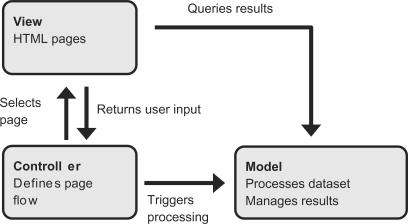
The architecture of MAGMA follows the Model-View-Controller design pattern, and the MAGMA web application is structured accordingly, as shown in Figure 1. The View part defines the HTML pages that are presented to the user. Any user action on the web pages is processed by the Controller who triggers an action in the Model and determines which page is to be displayed subsequently. The values and results that are shown on the HTML pages are directly requested from the model. This functionality separation enforced by the Model-View-Controller pattern enables user interface designers, java developers and statisticians to work independently on the HTML pages, the java part and the statistical data analysis. For the implementation we used the JSF technology (http://java.sun.com/javaee/javaserverfaces, JSF), which provides a library for all user interface elements, and a Controller that is set up with a straight forward XML configuration file. JSF greatly simplifies the generation of web pages and completely takes care of the low level HTTP request processing and user input handling. The data processing is done with R and Bioconductor packages. The R functionality is made available to the java server via Rserve (http://stats.math.uni-augsburg.de/Rserve). The microarray data and processing results are stored in R workspaces. The MAGMA web application can run on any web server that includes a servlet container, e.g. Apache Tomcat (http://tomcat.apache.org/). It is platform independent and has been successfully tested to run under Linux and Windows operating systems.
Figure 1.
Structural representation of MAGMA's architecture. MAGMA implements the Model-View-Controller paradigm and has the HTML pages (View), the rules for the HTML page flow (Controller) and the actual data processing (Model) decoupled. This allows for independent modification or extension of MAGMA.
MAGMA furthermore has an exception-handling mechanism, which provides feedback to users if inappropriate data or settings have been submitted. On the one hand, every input is syntactically validated at the time of submission, and a notice is displayed if invalid input is encountered. On the other hand, if unexpected results occur while processing the data, the processing step is aborted and an error page is displayed, explaining to the user the issue encountered, and providing suggestions how to resolve this.
PROGRAM DESCRIPTION
MAGMA provides for each user a separate workspace for storing and analyzing microarray data. The set of hybridization data files of a microarray study and their analysis are grouped together as an experiment within MAGMA. A typical analysis comprises the four processing steps upload, annotation, normalization and statistical analysis which are described in more detail subsequently. Users are intuitively guided through these four steps by the system. MAGMA has a navigation box (Figure 2) where the completed steps of an analysis are represented as distinct folders. Selecting a step icon links to the corresponding result page where the result box is shown in the lower area. Further processing steps can be run by selecting a suitable step from the Next box. The Manage experiments link is used to switch between different experiments. Info icons on all pages provide explanations for the individual boxes. For all input fields and links, short tool tips show up upon mouse over.
Figure 2.
MAGMA's result navigation panel. For each analyzed experiment, the results are organized in an explorer-like hierarchy. Each folder of the tree represents a single processing step and clicking on it, displays the corresponding result in the lower part of the page. More processing steps can be added after the current processing step by selecting a step from the Next box.
Each result box has in its title a link to the R-Script that generated the result. If R and Bioconductor are installed locally, this R-script can be pasted without any modification in the local R environment, and will then locally regenerate exactly the same processing results.
Upload
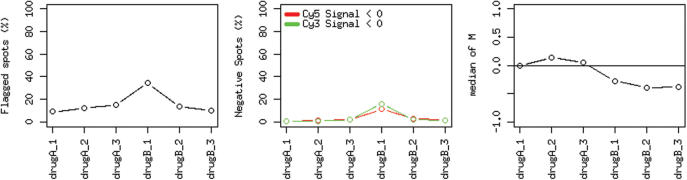
As input, MAGMA requires data files with raw and processed hybridization intensities from two-channel hybridization experiments. Currently, data files generated by the microarray image quantification applications Axon GenePix, Microdiscovery GeneSpotter and Agilent Feature Extraction software are supported. Further formats will be added on demand. All data files that should be analyzed together have to be packed into a single zip file. In the upload step MAGMA parses the files, computes diagnostic statistics from the extracted values and generates graphs that can be used as a first visual quality control. In particular, MAGMA computes the percentage of negative probes and the percentage of flagged (low-quality signal) probes. This allows the user to determine whether the background intensity and the noise of all hybridizations were acceptable. Additionally, the reported median log ratio of the red and the green intensities indicate for each file whether both channels are well balanced. These statistics are also displayed as graphs (Figure 3) and allow for the easy identification of outliers.
Figure 3.
Graphs visualizing diagnostic statistics. In the example, the percentage of flagged and negative probes as well as the median log ratio is shown for six uploaded arrays. These overview plots allow for the identification of outliers as well as general trends.
Annotation
In this step, the user should give short and accurate terms for the experimental conditions of the samples and assign them to the red and green channels of the data files. This is an important step, because the subsequent statistical analysis will use this information to group the data. The boxplots created in this step show whether the signal range within and across experimental conditions is consistent.
Normalization
The normalization step is optional, since MAGMA reads data that was already background corrected and normalized by the respective image quantification software. If this pre-normalization was not satisfying, the user can perform an optional background correction and a normalization using a set of methods provided by the Bioconductor package limma. After normalization, the MA-plots are recomputed so that the performance of the normalization can be evaluated visually.
Statistical analysis
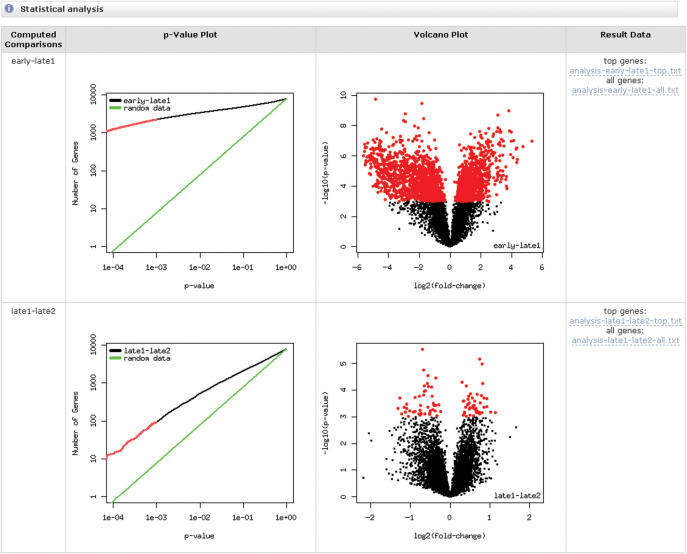
Finally, the statistical analysis generates a list of differentially expressed genes between any two of the experimental conditions defined in the annotation step. This list is computed again with the limma package which fits a linear model to the data. The limma package was chosen because it is the only tool that allows for the analysis of all common experimental designs (reference, circular, dye swap) for two-channel data with a simple consistent interface. Corrections for multiple testing can be selected as an advanced option and are computed using Bioconductor's multtest package. The result is displayed as a P-value plot that shows the number of significant genes as a function of the P-value threshold (Figure 4). The genes selected by the current threshold are highlighted in red. For a comparison, the number of significant genes that would be detected in random data is shown in green. The second plot is a volcano plot that shows the negative logarithm of the significance as a function of the logarithm of the fold change (Figure 4). These two plots show the amount of differential expression and the number of genes affected. The result table itself is available as a link and can be saved to any spreadsheet program.
Figure 4.
Result of a statistical analysis that included two comparisons. Each comparison is shown as a row with two plots. The first plot shows the number of significantly differentially expressed genes (red) selected using the current P-value threshold and compares it to results expected from random data. The second figure shows a Volcano plot displaying the P-values and fold changes of all genes with significant genes highlighted in red. The results can be retrieved in tabular form from the links in the rightmost column.
DISCUSSION
MAGMA combines Bioconductor's powerful statistical microarray analysis algorithms with state-of-the-art web application technology and offers the user the straightforward computation of differentially expressed genes with his web browser. While MAGMA's processing algorithms compare to those of other existing web applications, it stands out through its simple and intuitive usage and its comprehensive exception handling system, and therefore specifically aims at allowing novice users to analyze their data without prior training. MAGMA automatically generates R-scripts that guarantee the reproducibility of the generated results and thus allows advanced users to further extend the analysis. To our knowledge, it is the only web application where the generated R-script can be pasted without any modification into a local R/Bioconductor environment in order to reproduce all microarray data analysis results locally.
On the technical side, MAGMA demonstrates that modern software technologies, like the JSF framework, can be successfully applied even in small and concise academic projects and leads to well-structured solutions. The benefit of the chosen Java-based approach is several-fold:
Java is platform independent and runs on all major operating systems.
JSF technology comes with a comprehensive set of standard functionalities for web applications and simplifies the user interface development.
With Eclipse (www.eclipse.org), a freely available prime-class integrated development environment exists that supports collaborative software development, automatic code generation, instant syntax checking, source code versioning and integrated testing and deployment of web applications.
Java's object orientation intrinsically suggests the implementation of modular software packages that are easy to maintain and extend.
The MAGMA framework has already proven its flexibility and extensibility, as we have reused it to implement a dedicated web-based processing pipeline for the microarray data generated by the EuReGene project (www.euregene.org), with one person working less than a week on it.
ACKNOWLEDGEMENTS
We would like to thank for the critical feedback of many users in the beginning of the project. We thank further Christian Ahrens, Andrea Patrignani, Ulrich Wagner and Marzanna Künzli for discussions and comments on the manuscript. Many thanks also go to Snowflake Productions, Switzerland for the nice design of the web pages. Funding to pay the Open Access publication charges for this article was provided by the Functional Genomics Center Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. CARMAweb: comprehensive R- and Bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argraves GL, Jani S, Barth JL, Argraves WS. ArrayQuest: a web resource for the analysis of DNA microarray data. BMC Bioinformatics. 2005;6:287. doi: 10.1186/1471-2105-6-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapushesky M, Kemmeren P, Culhane AC, Durinck S, Ihmels J, Körner Ch, Kull M, Torrente A, Sarkans U, et al. Expression Profiler: next generation—an online platform for analysis of microarray data. Nucleic Acids Res. 2004;32:W465–W470. doi: 10.1093/nar/gkh470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luscombe NM, Royce TE, Bertone P, Echols N, Horak ChE, Chang JT, Snyder M, Gerstein M. ExpressYourself: a modular platform for processing and visualizing microarray data. Nucleic Acids Res. 2003;31:3477–3482. doi: 10.1093/nar/gkg628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Psarros M, Heber S, Sick M, Thoppae G, Harshman K, Sick B. RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res. 2005;32:W638–W643. doi: 10.1093/nar/gki490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hokamp K, Roche FM, Acab M, Rousseau M-E, Kuo B, Goode D, Aeschlimann D, Bryan J, Babiuk LA, et al. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res. 2004;32:W457–W459. doi: 10.1093/nar/gkh446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudsen S, Workman C, Sicheritz-Poten T, Friis C. GenePublisher: automated analysis of DNA microarray data. Nucleic Acids Res. 2003;31:3471–3476. doi: 10.1093/nar/gkg629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colantuoni C, Henry G, Zeger S, Pevsner J. SNOMAD (Standardization and Normalization of MicroArray Data): web accessible gene expression data analysis. Bioinformatics. 2002;18:1540–1541. doi: 10.1093/bioinformatics/18.11.1540. [DOI] [PubMed] [Google Scholar]
- 10.Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia X, McClelland M, Wang Y. WebArray: an online platform for microarray data analysis. BMC Bioinformatics. 2005;6:306. doi: 10.1186/1471-2105-6-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romualdi C, Vitulo N, Favero MD, Lanfranchi G. MIDAW: a web tool for statistical analysis of microarray data. Nucleic Acids Res. 2005;33:W644–W649. doi: 10.1093/nar/gki497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. NY: Springer; 2005. [Google Scholar]