Abstract
To distinguish the real pre-miRNAs from other hairpin sequences with similar stem-loops (pseudo pre-miRNAs), a hybrid feature which consists of local contiguous structure-sequence composition, minimum of free energy (MFE) of the secondary structure and P-value of randomization test is used. Besides, a novel machine-learning algorithm, random forest (RF), is introduced. The results suggest that our method predicts at 98.21% specificity and 95.09% sensitivity. When compared with the previous study, Triplet-SVM-classifier, our RF method was nearly 10% greater in total accuracy. Further analysis indicated that the improvement was due to both the combined features and the RF algorithm. The MiPred web server is available at http://www.bioinf.seu.edu.cn/miRNA/. Given a sequence, MiPred decides whether it is a pre-miRNA-like hairpin sequence or not. If the sequence is a pre-miRNA-like hairpin, the RF classifier will predict whether it is a real pre-miRNA or a pseudo one.
INTRODUCTION
MicroRNAs (miRNAs) are non-coding RNAs that can play important roles in gene regulation by targeting mRNAs for cleavage or translational repression (1,2). The miRNAs are transcribed as long primary miRNAs, which are processed into 60–70 nt miRNA precursors (pre-miRNAs) by nuclear RNase III Drosha (3). The pre-miRNAs are then cleaved into ∼22 nt mature miRNAs (4).
Because it is difficult to systematically detect miRNAs from a genome by existing experiment techniques, computational methods play important roles in the identification of miRNAs. It has been reported that miRNA genes are conserved in the primary sequences and secondary structures (2,5). Thus the comparative genomics-based methods were adopted to find novel miRNAs in specific animals and plants. MiRscan relies on the observation that the known miRNAs are derived from phylogenetically conserved stem-loop precursor RNAs with characteristic features (6,7). It successfully predicted hundreds of miRNAs in nematodes and human with a high sensitivity. The miRseeker (8) was developed for predicting miRNAs in insects, whereas MIRcheck (9) and MIRFINDER (10) were applied in plants. The miRAlign (11) aligns the secondary structure of pre-miRNAs to detect miRNAs.
Although those comparative genomics-based methods provided important techniques to predict new miRNAs, it is unable to identify novel miRNAs for which there are no known close homologies either due to the limitation of the data or due to the possible evolution of miRNAs. Furthermore, for a species that does not have a closely related species sequenced, its miRNAs cannot be studied with the comparative genomics approaches (12). So it is in high demand for ab initio prediction methods of miRNAs. Several studies show that many miRNAs are transcribed as polycistronic transcripts which are several kb long (13). Therefore, the genomic regions around the loci of known miRNAs appear particularly promising for discovering additional miRNAs. Sewer et al. (14) proposed a support vector machine (SVM) approach to identify the clustered miRNAs which were around already known miRNAs. Nam and co-workers constructed a highly specific probabilistic model (HMM) to search for distant homologs of miRNA families (15,16). They also showed that the integration of sequential and structural characteristics could improve the performance of a predictor in identifying clustered, non-clustered, conserved and non-conserved miRNAs (15,16). Yousef et al. (17) used a Naïve Bayes classifier along with the integration of data from multiple species to predict miRNA genes. The results indicate that by integrating data from multiple species, the model can be more likely to be applicable to a variety of genomes. As is suggested by Helvik et al. (18), the miRNA gene prediction methods can also be improved by reliable predictions of Drosha-processing sites (18). Recently, genome-wide surveys for non-coding RNAs have provided evidence for tens of thousands of previous undescribed evolutionary conserved RNAs with distinctive secondary structures (19). In contrast to other miRNA detection methods which directly search a genome or genomes, RNAmicro (20) is designed to classify the raw results of large-scale comparative genomics surveys for putative RNAs that are conserved in both sequence and secondary structure.
Although almost all pre-miRNAs have the characteristic of stem-loop hairpin structures (1,6,7,11), a large amount of pre-miRNA-like hairpins can be folded in many genomes. It is still a challenge to distinguish the real pre-miRNAs from other hairpin sequences with similar stem-loops (pseudo pre-miRNAs). Xue et al. (21) proposed an SVM-based method for classification of real and pseudo pre-miRNAs. The local contiguous structure-sequence composition feature was used. A Perl package (Linux system only), Triplet-SVM-classifier, was provided. However, as was indicated that pre-miRNAs, unlike tRNAs and rRNAs, had lower folding free energies than random sequences (22), the thermodynamics-related features might improve the prediction performance.
In this article, in order to achieve higher performance of distinguishing the real pre-miRNAs from the pseudo ones, a hybrid feature by incorporating the local contiguous structure-sequence composition, the minimum of free energy (MFE) of the secondary structure and the P-value of randomization test was used. Besides, a novel machine-learning algorithm, random forest (RF), was introduced. The results indicated that our method significantly outperformed the Triplet-SVM-classifier. Furthermore, an alternative classifier which used the SVM with the hybrid feature was also compared with the Triplet-SVM-classifier and our RF-based method. The results showed that the alternative classifier outperformed the Triplet-SVM-classifier, but it underperformed our RF method. It indicated that both the RF algorithm and the hybrid feature contributed to the prediction improvement.
A web server (MiPred) is available at http://www.bioinf.seu.edu.cn/miRNA/. Given a sequence, MiPred decides whether it is a pre-miRNA-like hairpin sequence or not. If the sequence is a pre-miRNA-like hairpin, the RF classifier will predict whether it is a real pre-miRNA or a pseudo one.
MATERIALS AND METHODS
Data sets
Human real pre-miRNAs
Human pre-miRNAs are downloaded from the miRNA registry database (23) in August 2006 (release 8.2), which contains 462 reported pre-miRNA entries from Homo sapiens. Only the pre-miRNAs whose secondary structures do not contain multiple loops are considered, which gives us 426 pre-miRNAs, covering >92% of all the reported human pre-miRNAs.
Human pseudo pre-miRNAs
The human pseudo pre-miRNAs were obtained from http://bioinfo.au.tsinghua.edu.cn/mirnasvm/, which contained 8494 pre-miRNA-like hairpins (21). As all reported miRNAs are located in the un-translated regions or intergenic regions, the data set was collected from the protein coding regions. The criteria for selecting the pseudo pre-miRNAs from the segments are: (i) The sequence length ranges from 51 nt to 137 nt; (ii) minimum of 18 base pairings on the stem of the hairpin structure (included the GU wobble pairs); (iii) maximum of −15 kal/mol free energy of the secondary structure; (iv) no multiple loops. The criteria ensure that the extracted pseudo pre-miRNAs are similar to real pre-miRNAs according to the widely accepted characteristics. (The thresholds 51, 137, 18 and −15 are the shortest length, the longest length, the lowest number of base pairings and the highest free energy among all the genuine human pre-miRNAs, respectively.)
Training data set and testing data set
We trained our RF prediction model on the same training data set of the Triplet-SVM-classifier (21), which contained 163 real pre-miRNAs and 168 pseudo pre-miRNAs. The testing data set comprised of the remaining 263 real pre-miRNAs not used in the training data set and the 265 pseudo pre-miRNAs randomly picked up from the human pseudo pre-miRNAs data set (samples already selected in the training data set were avoided).
The minimum of free energy (MFE) feature
The minimum of free energy (MFE) of the secondary structure was predicted by the Vienna RNA software package (24).
The local contiguous triplet structure composition
In the predicted secondary structure, there are only two statuses for each nucleotide, paired or unpaired, indicated by brackets ‘(‘or’)’ and dots ‘.’, respectively. The left bracket ‘(‘means the paired nucleotide is located near the 5′-end and can be paired with another nucleotide at the 3′-end, which is indicated as a right bracket ‘)’. Here, we do not distinguish these two situations and use ‘(’ for both situations. For any three adjacent nucleotides, there are eight (23) possible structure compositions: ‘(((’, ‘((.’, ‘(..’, ‘…’, ‘.((’, ‘..(’, ‘.(.’ and ‘(.(’. Considering the middle nucleotide among the three, there are 32 (4 × 8) possible structure-sequence combinations, which are denoted as ‘U(((’, ‘A((.’, etc. The local contiguous triplet structure composition is defined as the fraction of each triplet structure-sequence element in the appearance of the all possible triplet elements (21).
Dinucleotide shuffling
Dinucleotide shuffling is to shuffle a sequence while keeping the dinucleotide distribution (or frequencies) constant. It has been demonstrated that random RNA must be generated with the same dinucleotide frequency, for any valid conclusions to be drawn (25). An implementation of the algorithm as described by Workman and Krogh (25) was used.
The P-value of randomization test feature
In order to determine if the MFE value is significantly different from that of random sequences, a Monte Carlo randomization test was used (22). The test can be summarized as follows:
Compute MFE of the secondary structure inferred from the original sequence.
Randomize the order of the nucleotides in the original sequence while keeping the dinucleotide distribution (or frequencies) constant. Then compute the MFE for the inferred structure based on the shuffled sequence.
Repeat step 2 a great number of times (1000) in order to build the distribution of MFE values.
If N is the number of iterations and R the number of randomized sequences that have a MFE value less or equal to the original value, then P-value is defined as:
Random forest
Random forest (RF) is a classifier consisting of an ensemble of tree-structured classifiers (26). RF takes advantage of two powerful machine-learning techniques: bagging (27) and random feature selection. In bagging, each tree is trained on a bootstrap sample of the training data, and predictions are made by majority vote of trees. RF is a further development of bagging. Instead of using all features, RF randomly selects a subset of features to split at each node when growing a tree. To assess the prediction performance of the random forest algorithm, RF performs a type of cross-validation in parallel with the training step by using the so-called out-of-bag (OOB) samples. Specifically, in the process of training, each tree is grown using a particular bootstrap sample. Since bootstrapping is sampling with replacement from the training data, some of the sequences will be ‘left out’ of the sample, while others will be repeated in the sample. The ‘left out’ sequences constitute the OOB sample. On average, each tree is grown using about 1 − e−1 ≅ 2/3 of the training sequences, leaving e−1 ≅ 1/3 as OOB. Because OOB sequences have not been used in the tree construction, one can use them to estimate the prediction performance (28). The RF algorithm was implemented by the randomForest R package (29).
Support vector machine
SVM is a supervized machine-learning technology based on statistical theory for data classification (30). SVM seeks an optimal hyperplane to separate two classes of samples. It uses kernel functions to map original data to a feature space of higher dimensions and locate an optimal separating hyperplane there. The SVM algorithm was implemented by the e1071 (version 1.5–12) R package (31).
Prediction system assessment
For a prediction problem, a classifier can classify an individual instance into the following four categories: false positive (FP), true positive (TP), false negative (FN) and true negative (TN). The total prediction accuracy (ACC), Specificity (Sp), Sensitivity (Se) and Mathew's correlation coefficient (MCC) (32) for assessment of the prediction system are given by
| 1 |
| 2 |
| 3 |
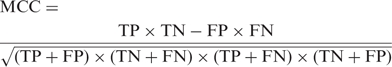 |
4 |
RESULTS AND DISCUSSION
The RF prediction performance using combined features
The prediction results using the RF prediction modules with various features are shown in Table 1. The performance was assessed by OOB estimation on the training data set. The local contiguous triplet structure composition-based RF classifier was able to predict with 88.21% total accuracy and 0.77 MCC value. If we combined the MFE of the secondary structure or P-value feature with the local contiguous triplet structure composition feature, the prediction performance significantly increased (MFE + local contiguous triplet structure composition: 93.35% ACC and 0.87 MCC; P-value + local contiguous triplet structure composition: 96.07% ACC and 0.92 MCC). It indicated that the MFE and the P-value used in our article were two key attributes which discriminated the real microRNA precursors from the pseudo ones. The combination of all features achieved the best performance with 96.68% ACC and 0.94 MCC value. These results indicate that a combined feature vector is capable of extracting more information about a primary sequence and obtaining a better prediction performance.
Table 1.
The performance of RF prediction modules based on various features. The prediction system was assessed by OOB estimation on data set 1.
| Features | Sp (%) | Se (%) | ACC (%) | MCC |
|---|---|---|---|---|
| A | 90.48 | 85.89 | 88.21 | 0.77 |
| A + B | 95.24 | 91.41 | 93.35 | 0.87 |
| A + C | 97.62 | 94.47 | 96.07 | 0.92 |
| A + B + C | 98.21 | 95.09 | 96.68 | 0.94 |
A: local contiguous triplet structure composition;
B: Minimum of free energy (MFE) of the secondary structure;
C: P-value.
Estimating and ranking the feature importance
Decision tree is known for its ability to select ‘important’ ones from many features and ignore (often irrelevant) others. In addition, decision tree gives an explicit model describing the relationship between features and predictions, thus easing model interpretation. Random forest, as an ensemble of trees, inherits the ability to select ‘important’ features. A measure of how each feature contributes to the prediction performance of random forest can be calculated in the course of training. When a feature that contributes to prediction performance is ‘noised up’ (e.g. replaced with random noise), the performance of the prediction is noticeably degraded. On the other hand, if a feature is irrelevant, ‘noising’ it up should have little effect on the performance. Thus, we can estimate the relative importance of features according to the following procedure (28). For each tree, the prediction accuracy on the OOB portion of the data is recorded. Then the same is done after permuting each predictor variable. The difference between the two accuracies are then averaged over all trees, and normalized by the standard error. Then the MDA implies the relative importance of each feature. As shown in Table 2, P-value, MFE and ‘C …, U(((, A(((’ composition are the top five features which determined the pre-miRNA-like hairpins to be the real or pseudo ones. ‘A((., U.((, C(.(’ triplet elements seemed to have no discriminative power in the RF classifier.
Table 2.
Estimating and ranking the relative importance of the features
| Rank | Features | Mean decrease accuracy (%) |
|---|---|---|
| 1 | P-value | 15.80 |
| 2 | MFE | 5.48 |
| 3 | C … | 2.04 |
| 4 | U((( | 2.00 |
| 5 | A((( | 1.49 |
| 6 | A … | 0.83 |
| 7 | G … | 0.76 |
| 8 | U..( | 0.43 |
| 9 | G.(( | 0.34 |
| 10 | A(.. | 0.31 |
| 11 | C((. | 0.31 |
| 12 | G..( | 0.29 |
| 13 | G(.. | 0.29 |
| 14 | U((. | 0.27 |
| 15 | U … | 0.26 |
| 16 | U(.( | 0.24 |
| 17 | G((( | 0.23 |
| 18 | C((( | 0.20 |
| 19 | A..( | 0.20 |
| 20 | U(.. | 0.19 |
| 21 | C(.. | 0.14 |
| 22 | U.(. | 0.14 |
| 23 | G((. | 0.09 |
| 24 | C.(. | 0.09 |
| 25 | A(.( | 0.08 |
| 26 | C..( | 0.08 |
| 27 | C.(( | 0.07 |
| 28 | A.(. | 0.07 |
| 29 | G(.( | 0.06 |
| 30 | A.(( | 0.03 |
| 31 | G.(. | 0.02 |
| 32 | A((. | 0.00 |
| 33 | U.(( | 0.00 |
| 34 | C(.( | 0.00 |
Comparison with other methods
The performance of our RF method was compared with the existing method, Triplet-SVM-classifier (21), which was also trained on the same training data set. The results were obtained by an independent data set test. The testing data set contained 263 real pre-miRNAs and 265 pseudo pre-miRNAs. As was shown in Table 3, the results demonstrated that the total prediction accuracy of our RF method was nearly 10% greater than the Triplet-SVM-classifier. An alternative classifier which used the SVM method with the combined features was also compared with the Triplet-SVM-classifier and our RF-based method. The results showed that the alternative classifier significantly outperformed the Triplet-SVM-classifier, but it slightly underperformed the RF-based method. To further compare the RF algorithm with the SVM algorithm, we combined the training data set and the testing data set, and then randomly divided it into two portions of approximately equal size. We used one portion for training and the other for evaluating the prediction performance. The process was repeated 20 times. The results showed that the mean accuracy of the RF method is 93.49% which is higher than that of the SVM method (92.94%). A pairwise t-test was also implemented. The P-value was 0.003, which indicated that the performance between the RF and the SVM was significantly different. Thus, we conclude that both the RF algorithm and the hybrid feature contribute to the prediction improvement.
Table 3.
Comparison with the existing method and the competing method. All the algorithms are trained on the same training data set and tested on the same testing data set
| Methods | Sp (%) | Se (%) | ACC (%) | MCC |
|---|---|---|---|---|
| RF | 93.21 | 89.35 | 91.29 | 0.826 |
| SVM | 90.94 | 87.83 | 89.39 | 0.788 |
| Triplet-SVM-classifier | 88.30 | 79.47 | 83.90 | 0.681 |
RF: An RF-based method with ‘P-value + MFE + local contiguous triplet structure composition’ features;
SVM: An SVM-based method with ‘P-value + MFE + local contiguous triplet structure composition’ features;
Triplet-SVM-classifier: An SVM-based method with Local contiguous triplet structure composition features.
The miR-abela (14) used an ab initio prediction method to identify pre-miRNA candidates. The prediction process follows the same strategy as our method: (i) initial screen of miRNA genes; (ii) using machine-learning methods to distinguish the real miRNAs from the pseudo ones. However, the performance of a machine-learning method depends on the sensitivity and specificity of the initial screen. The miR-abela and our method use different initial screen rules and different pseudo sample definitions (negative samples of machine-learning algorithm in training datasets). Thus, it is a challenge to directly compare them in a given data set. The miRBase (http://microrna.sanger.ac.uk/) deposits several newly found pre-miRNAs from August 2006 (release 8.2) to February 2007 (release 9.1). We use those sequences (www.bioinf.seu.edu.cn/miRNA/dataset1.htm) to compare our method with miR-abela. As shown in Table 4, our method (MiPred) successfully predicts the newly found pre-miRNAs with 100% accuracy while the total accuracy of miR-abela is only 46.34%. Furthermore, another independent data set: Human cytomegalovirus miRNAs (miRBase: release 9.1), which consists of 11 pre-miRNAs (www.bioinf.seu.edu.cn/miRNA/dataset2.htm), is used to compare our MiPred with miR-abela in identifying long un-related pre-miRNAs. The results show that MiPred still obtains 100% accuracy while the total accuracy of miR-abela is only 27.27% (3/11, 3: the number of sequences which is correct predicted; 11: the total number of sequences).
Table 4.
Prediction accuracy on an independent data set test
| Species | Accuracy | |
|---|---|---|
| MiPred | miR-abela | |
| Homo sapiens | 100% (2/2) | 0% (0/2) |
| Rattus norvegicus | 100% (1/1) | 100% (1/1) |
| Caenorhabditis elegans | 100% (18/18) | 33.3%(6/18) |
| Mus musculus | 100% (4/4) | 25% (1/4) |
| Caenorhabditis briggsae | 100% (16/16) | 68.75% (11/16) |
| Total | 100% (41/41) | 46.34% (19/41) |
ProMiR II (15) is a web server that search for potential miRNAs in a given sequence or in its vicinity. It provides three programs: ProMiR-v (search for potential miRNAs in the Vicinity of known miRNAs), ProMiR-c (search for potential miRNAs in the vicinity of a Candidate) and ProMiR-g (predict miRNAs in a long sequence, a Generalized version of ProMiR). There are 13 known miRNAs in Chromosome II of Homo sapiens: has-mir-558, has-mir-559, has-mir-217, has-mir-216, has-mir-560, has-mir-128a, has-mir-10b, has-mir-561, has-mir-26b, has-mir-375, has-mir-153-1, has-mir-562 and has-mir-149. We use ProMiR-v to search for potential miRNAs in the vicinity of these sequences. The default parameters are used. The results show that there are seven known pre-miRNAs and 21 pre-miRNA candidates (computationally predicted as pre-miRNAs). Then we take those 21 candidates (www.bioinf.seu.edu.cn/miRNA/dataset3.htm) to run in our MiPred. The results show that the miRNA-candidates which are predicted as real pre-miRNAs in ProMiR-v are also predicted as real ones in MiPred. In addition, we randomly extract ten sequences (each with length 10k nt) from Chromosome III–VIII of Homo sapiens and use ProMiR-g to search for potential pre-miRNAs from those sequences. It detects three sub-sequences as potential pre-miRNAs. We test those three sequences using MiPred and find that our prediction results are consensus with those of ProMiR-g: the miRNA-candidates which are predicted as real pre-miRNAs in ProMiR-g are also predicted as real ones in MiPred. Thus, we conclude that although the working principle is different between MiPred and ProMiR II, the two web servers have a very consensus result.
To detect miRNA-candidates from computational approach, it is vital important to use different web servers. Thus, conflicting results can be noted and evaluated by users. MiPred will provide a useful tool to detect and evaluate miRNA-candidates.
Server description
MiPred is available at http://www.bioinf.seu.edu.cn/miRNA/. All the CGI scripts of the method were written in Perl 5.8.8 and the interface was designed using HTML. The RF algorithm was implemented by the randomForest R package (29) and the MFE was predicted by the Vienna RNA software package (24). In the current system, the training data set is the same as that of the Triplet-SVM-classifier, which contains 163 real pre-miRNAs and 168 pseudo pre-miRNAs.
Users can enter a RNA sequence (uppercase or lowercase) in one of four formats (FASTA, GCG, GeneBank and EMBL). All non-standard characters except the four nucleotide bases adenine, guanine, cytosine and uracil will be ignored from the sequence.
Given a sequence, MiPred decides whether it is a pre-miRNA-like hairpin sequence or not. If the sequence is a pre-miRNA-like hairpin, the RF classifier will predict whether it is a real pre-miRNA or a pseudo one. Besides, the prediction confidence of the RF classifier is also provided. Here the prediction confidence is defined as the fraction of positive votes for the predicted real pre-miRNA or the fraction of negative votes for the predicted pseudo pre-miRNA. An output example is shown in Figure 1.
Figure 1.
Prediction results of a query RNA sequence.
CONCLUSIONS
We have devised an RF-based method for classification of the real pre-miRNAs and the pseudo pre-miRNAs using a hybrid feature. We compared our method with the existing method, Triplet-SVM-classifier (21), which was also trained on the same training data set. The results demonstrated that the total prediction accuracy of our RF method was nearly 10% greater than the Triplet-SVM-classifier. Further analysis indicated that the improvement was due to both the hybrid feature and the RF algorithm. We also compared our method with miR-abela and ProMiR II using independent data sets test. The results indicated that our method significantly outperformed miR-abela but had a very consensus result with ProMiR II. A web server MiPred was developed. Given a sequence, MiPred decides whether it is a pre-miRNA-like hairpin sequence or not. If the sequence is a pre-miRNA-like hairpin, the RF-based classifier will predict whether it is a real pre-miRNA or a pseudo one.
Scanning the genome, there could be numerous amounts of sequence segments that can be folded into pre-miRNA like hairpins. The successful ab initio classification of real and pseudo pre-miRNAs opens a new approach for discovering new miRNAs.
ACKNOWLEDGEMENTS
The work is supported by National Natural Science Foundation of China (Project No. 60121101).Funding to pay Open access publication charges for this article was provided by the National Natural Science Foundation of China (No. 60121101).
Conflict of interest statement. None declared.
REFERENCES
- 1.Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends. Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Stricker HM, Gou D, Liu L. MicroRNA: past and present. Front Biosci. 2007;12:2316–2329. doi: 10.2741/2234. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Gesellchen V, Boutros M. Managing the genome: microRNAs in Drosophila. Differentiation. 2004;72:74–80. doi: 10.1111/j.1432-0436.2004.07202003.x. [DOI] [PubMed] [Google Scholar]
- 6.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 8.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhang J, Li F, Gu J, He T, Zhang X, Li Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- 12.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 13.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sewer A, Paul N, Landgraf P, Aravin A, Pfeffer S, Brownstein MJ, Tuschl T, van Nimwegen E, Zavolan M. Identification of clustered microRNAs using an ab initio prediction method. BMC Bioinformatics. 2005;6:267. doi: 10.1186/1471-2105-6-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam JW, Kim J, Kim SK, Zhang BT. ProMiR II: a web server for the probabilistic prediction of clustered, nonclustered, conserved and nonconserved microRNAs. Nucleic Acids Res. 2006;34:W455–W458. doi: 10.1093/nar/gkl321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nam JW, Shin KR, Han J, Lee Y, Kim VN, Zhang BT. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 2005;33:3570–3581. doi: 10.1093/nar/gki668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yousef M, Nebozhyn M, Shatkay H, Kanterakis S, Showe LC, Showe MK. Combining multi-species genomic data for microRNA identification using a Naive Bayes classifier. Bioinformatics. 2006;22:1325–1334. doi: 10.1093/bioinformatics/btl094. [DOI] [PubMed] [Google Scholar]
- 18.Helvik SA, Snove O, Jr, Saetrom P. Reliable prediction of Drosha processing sites improves microRNA gene prediction. Bioinformatics. 2007;23:142–149. doi: 10.1093/bioinformatics/btl570. [DOI] [PubMed] [Google Scholar]
- 19.Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hertel J, Stadler PF. Hairpins in a Haystack: recognizing microRNA precursors in comparative genomics data. Bioinformatics. 2006;22:e197–e202. doi: 10.1093/bioinformatics/btl257. [DOI] [PubMed] [Google Scholar]
- 21.Xue C, Li F, He T, Liu GP, Li Y, Zhang X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310. doi: 10.1186/1471-2105-6-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Workman C, Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breiman L. Random forest. Mach. Learning. 2001;45:5–32. [Google Scholar]
- 27.Breiman L. Bagging predictors. Mach. Learning. 1996;24:123–140. [Google Scholar]
- 28.Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 29.Liaw A, Wiener M. Classification and Regression by randomForest. R. News. 2002;2:18–22. [Google Scholar]
- 30.Vapnik V. Statistical Learning Theory. NY: Wiley; 1998. [DOI] [PubMed] [Google Scholar]
- 31.Dimitriadou E, Hornik K, Leisch F, Meyer D, Weingessel A. e1071: Misc Functions of the Department of Statistics (e1071) 2006. TU Wien. R package, Version 1.5–12. Available from http://cran.R-project.org.
- 32.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta. 1975;405:442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]



