Abstract
The current Protein Data Bank (PDB) contains about 40 000 protein structures with approximately half a million incorrect atom positions resulting from erroneously assigned asparagine (Asn) and glutamine (Gln) rotamers. These errors affect applications in protein structure analysis, modeling and docking and therefore the detection, correction and prevention of such errors is highly desirable. We present NQ-Flipper, a web service based on mean force potentials to automatically detect and correct erroneous Asn and Gln rotamers. The service accepts protein structure files formatted in PDB style or PDB codes. For an Asn/Gln side-chain amide NQ-Flipper computes the total interaction energy with the surrounding atoms as the sum of pairwise atom–atom interaction energies. The energy difference between the original and the alternative rotamers identifies the correct configuration of the amide group. The web service lists the interaction energies of all Asn/Gln residues found in a PDB file and shows the structure and offending residues in an interactive 3D viewer. The corrected protein structure is available for download in various compression formats. The web service is accessible at http://flipper.services.came.sbg.ac.at
INTRODUCTION
The side-chains of the amino acids asparagine (Asn) and glutamine (Gln) terminate by an amide group. The amide group may form several hydrogen bonds with the surrounding atoms and thus frequently plays an important role in protein structure stability and in intermolecular interactions (1–8). A single amide group may form up to four hydrogen bonds, two donated by the nitrogen and two accepted by the oxygen. Hence, an incorrect configuration generally results in highly unfavorable interaction energies and in view of the refinement protocols used in X-ray analysis, which frequently employ energy calculations, the occurrence of errors may seem unlikely. Nevertheless, the average error rate found in the current Protein Data Bank (PDB) (9) is of the order of 20% (10–14). The high error rate arises from limitations encountered in X-ray analysis. The amide nitrogen and oxygen atoms have similar electron densities and are indistinguishable in electron density maps at moderate and low resolutions and it is generally thought that structures of higher resolution (1.5 Å or better) are largely error free. From this point of view the Asn/Gln rotamer problem seems to be a peculiarity of X-ray analysis. However, a similar error rate is found for solution structures of proteins determined by NMR (10).
Presently two web services provide information on Asn/Gln rotamers. The PDBREPORT database/WHATIF server (15) presents a static view on a plethora of automatically generated quality scores. Among these is a listing of incorrect Asn/Gln rotamers. The WHATIF server displays information but does not provide a mechanism to correct erroneous Asn/Gln rotamers. The MolProbity (16) server is dedicated to protein structure analysis and offers a variety of interactive tools to validate either an uploaded coordinate file or a structure from PDB. One special subtask is the validation and correction of Asn/Gln rotamers by adding hydrogens and picking the rotamer with lesser steric clashes.
Here we present NQ-Flipper, a web service for interactive validation and correction of Asn/Gln amide rotamers. The server operates in three steps. A protein coordinate file in PDB format is uploaded and scores based on potentials of mean force (10, 17) are computed. In the second step, the results are displayed in a table where incorrect Asn/Gln rotamers are marked and the respective structure is shown in an interactive 3D Java applet (http://www.jmol.org) where any offending Asn/Gln residues are highlighted. In the third step a coordinate file with corrected atom positions is produced, which may be downloaded from the server in various compression formats. If desired any changes suggested by the server may be edited and overruled by the user.
METHODS
NQ-Flipper employs knowledge-based potentials of mean force (17–20) derived from complete crystal structures. The atom types consist of all non-hydrogen atoms of standard amino acids found in the ATOM records of PDB files. However, the backbone atoms N, Cα, C and O of individual amino acids are treated as distinct atom types. Potentials of mean force do not require hydrogen atom positions. This is an advantage over potential functions where hydrogen atoms are needed for energy evaluation. Since hydrogen atoms are generally not visible in electron densities, their coordinates have to be inferred from the attached heavy atoms. Therefore, derived hydrogen positions do not provide new information and since such positions are frequently ambiguous they may actually falsify experimental data. Rare atom types frequently found in ligands and non-standard groups as well as water molecules whose positions are frequently unreliable are presently omitted from NQ-Flipper calculations.
The mean force potentials are derived from a non-redundant database of protein structures and are refined to a stable self-consistent set of potentials by repeated rotamer correction and potential recompilation (10). For any two atoms k and i of atom types ak and bi separated by a distance rki the mean force potential is given by ε (ak,bi,rki). The interaction at rki is attractive if ε (ak,bi,rki) < 0 and repulsive if ε (ak,bi,rki) > 0. We denote by R1 the original rotamer found in crystal structures and by N1 and O1 the associated side-chain amide atoms. The total interaction energy is computed by
where k = N1 and l = O1 and the summation is over all atoms i. For the alternative rotamer R2 the nitrogen and oxygen atoms swap their position and the respective energy ε (R2) is computed analogously with k = N2 and l = O2. The energy difference Δ ε: = ε (R1)-ε (R2) serves as a score and indicates the choice of the correct rotamer. The probability or expected relative occupancy is derived from the energy difference. A probability close to unity indicates that R1, the configuration found in the crystal structure, corresponds to the correct rotamer (11). Conversely, when this probability approaches zero the alternative rotamer R2 is highly favored and the configuration found in the crystal structure is incorrect. Probabilities between 0.1 and 0.9 indicate that both rotamers may be occupied and they most likely coexist in the crystal structure. A comparison with the reference data set of Word et al. results in an overall accuracy of 95.8% (10, 11).
WEB SERVER USAGE
The NQ-Flipper web service is a computational tool to validate and correct Asn/Gln rotamers in protein structures. Rotamers are reported with their associated Δε-score and erroneous rotamers are replaced by their corrected conformations resulting in a separate coordinate file available for download. NQ-Flipper has a small set of parameters that may be controlled and optionally changed by the user. Protein structures are entered at the main page along with a small number of parameters as described in the following paragraphs.
PDB code
A valid PDB four letter code specifies the protein structure to be processed. The repository of coordinate files maintained by NQ-Flipper is concurrent with PDB (9).
File name
Any coordinate file compliant with the PDB file format can be uploaded. When determining a new structure this allows rotamer validation at any stage of the protein crystal structure refinement process. Crystallographic symmetry is used to generate the complete crystal structures so that complete structures are used for rotamer assignment. Valid input file compression formats include gzip and unix compress.
Model number
Computations for coordinate files containing more than one model are restricted to a single model identified by this number.
Altloc indicator
Treatment of alternate location (altloc) indicators is similar to model numbers in that coordinates are retrieved for a certain altloc character only. The usage of altloc indicators is not clearly defined by PDB. In extreme cases all possible combinations of atoms with alternate locations may have to be considered. Depending on the particular PDB file this may result in an enormous number of possible combinations. A consistent treatment of alternate locations requires the submission of a complete model for each variant.
Threshold
A threshold v applied to Δε-scores is used to distinguish rotamers with single and multiple occupations. The larger the value of v the larger is the number of Asn/Gln amides considered to occupy both rotamers. The NQ-Flipper web page provides a statistical analysis of the agreement of NQ-Flipper results with a refined reference data set (12) as a function of the threshold v.
Rotate/Flip
Coordinates for the alternative rotamer atoms are derived by a 180° rotation about the Cβ - Cγ (Asn) and Cγ - Cδ (Gln) axis. This preserves bond angles and distances which is not guaranteed for the ‘Flip’ option where only atom identities are swapped. The latter option has been applied to correct Asn/Gln rotamers in the reference set (12). The option is offered here for comparison and the computation of the statistical analysis, but its use correcting Asn/Gln rotamers is strongly discouraged, since it results in improper covalent geometry of the amide atoms.
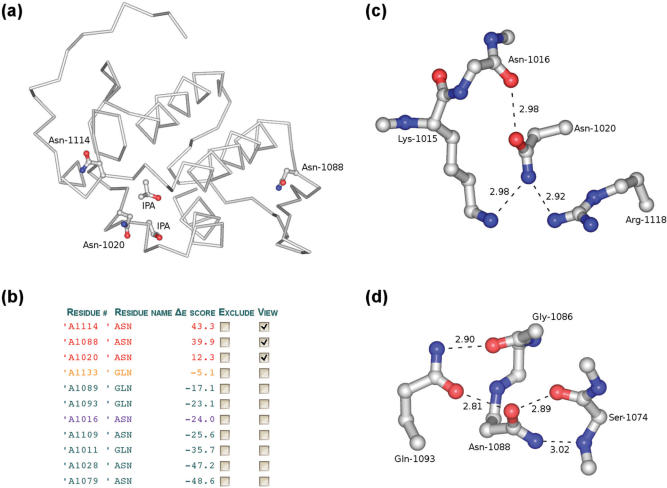
For a moderately sized protein the Asn/Gln rotamers are validated within seconds. The results are presented in tabular form listing all Asn/Gln residues sorted and color-coded by their associated Δε-score. Entries highlighted in red refer to rotamers with unfavorable interaction energies. An interactive 3D-viewer based on Jmol (http://www.jmol.org) displays the Cα backbone trace of the protein structure with side-chain atoms of incorrect rotamers rendered as spheres. Multiple occupancies (| Δ ε | < v) are indicated in orange and are left in their original conformation. Residues with side-chain amides closer than 8 Å to atom centers of non-standard groups are colored in blue. These are frequently residues participating in functional sites and therefore, they may be particularly important. The assignment produced by NQ-Flipper may be edited by the user. Corrected coordinate files can be downloaded in various compression formats. Figure 1 provides an example of acutohaemolysin, PDB code 1mc2 (alternate location indicator A) (21), a phospholipase A2 at a resolution of 0.85 Å directly solved by dual-space Shake-and-Bake refinement. The structure contains eleven Asn/Gln residues. Out of these NQ-Flipper flags three rotamers as erroneous based on disagreement with empirical statistics which is also in line with basic physico-chemical principles.
Figure 1.
NQ-Flipper analysis of Asn/Gln rotamers. We provide an example of the application of NQ-Flipper to acutohaemolysin (PDB code 1mc2) determined at 0.85 Å resolution (21). The PDB file contains a single chain (identifier A) of 122 amino acid residues. The molecule contains eleven Asn/Gln residues. Three of these residues are flagged as incorrect rotamers which is confirmed by a detailed examination based on simple physico-chemical principles. The structure is determined to a very high resolution where errors in rotamer configuration may seem unlikely. However, the result shown here is quite common. Generally protein structures, even at very high resolution, contain several incorrect rotamers. (a) The protein backbone C-alpha trace with unfavorable rotamers rendered as a ball-and-stick model. The crystal structure contains two heterogeneous groups termed IPA. (b) Table listing all Asn/Gln rotamers and their respective Δε-values as displayed on the NQ-Flipper web page. Coordinates were derived for alternate location indicator A. The software detects three Asn residues with unfavorable Δε-scores, i.e., rotamers with Δ ε > v where the threshold value is set to v = 6 (the default). These incorrect rotamers are highlighted in red and include Asn-1114 (Δ ε = + 43.3), Asn-1088 (Δ ε = + 39.9) and Asn-1020 (Δ ε = + 12.3). One residue, Gln-1133 (Δ ε = -5.1), is considered ‘ambiguous’ since its Δε-score is in the range where both rotamers are occupied with non-negligible probability (| Δ ε | < 6). Such residues of multiple occupancy are colored orange. All remaining Asn/Gln residues have Δε-scores below -6 indicating correct rotamers. Residue Asn-1016 (Δ ε =-24.0) shown in blue is in the vicinity of an isopropanol buffer molecule, IPA. The minimum distance between atoms from the IPA molecule and the respective amide group in Asn-1016 is 5.7 Å. The residue is colored blue to indicate that it is close to a heterogeneous group so that some important interactions may not be included in the score—although even in such cases a score greater than v generally indicates an incorrect rotamer. Figure 1c and d provide a detailed view of two of the incorrect rotamers in the conformation R1, as found in the PDB file, interacting with their respective chemical environment. The atoms are colored by atom type: carbon, gray; nitrogen, blue; oxygen, red. The dashed lines indicate distances in Å between atoms. (c) In the original conformation residue Asn-1020 (Δ ε = + 12.3) is unable to participate in a hydrogen bond network. In the alternative conformation R2 the amide nitrogen and oxygen atoms swap positions enabling hydrogen bond formation between the amide nitrogen and the carbonyl oxygen of Asn-1016, the amide oxygen and the side-chain nitrogen of Lys-1015 and a nitrogen of the guanidinium group of Arg-1118. Note that by flipping the amide group several ‘anti-hydrogen bonds’ of very high energy are replaced by genuine hydrogen bonds of very low energy. (d) Residue Asn-1088 (Δ ε = + 39.9), where the amide nitrogen and oxygen atoms have unfavorable interactions with the respective main chain nitrogen and oxygen atoms of Ser-1074. The amide oxygen of this residue additionally interacts unfavorably with Gln-1093, a correct rotamer since it forms a hydrogen bond to the carbonyl group of Gly-1086. Flipping Asn-1088 to rotamer R2 results in the formation of three hydrogen bonds. Figure a, c and d were generated using PyMOL (http://pymol.sourceforge.net).
CONCLUSION
The NQ-Flipper web service provides an interactive tool for the detection and correction of unfavorable Asn/Gln rotamers utilizing knowledge-based potentials of mean force derived from high resolution protein structures. Except for very large crystal structures the response time of the server is immediate (i.e. within seconds). The NQ-Flipper pages provide an easy to use and robust interface. Different colors relate Δε-scores to correct and incorrect rotamers, and to amides having multiple occupations. Any assignment made by the program can be edited by the user and the corrected coordinate file can be downloaded. The results obtained from NQ-Flipper largely agree with data on correct and incorrect Asn/Gln rotamers validated by experts (10, 11).
The software is available freely as a web service at http://flipper.services.came.sbg.ac.at. For the integration of NQ-Flipper with X-ray analysis or NMR refinement protocols, a stand-alone Linux version is available for download on the home page. Protein structures available from the PDB database are analyzed by entering the PDB four letter code, model number and alternate location indicator of the respective file. The service may also be accessed without entering data in the HTML form by directly supplying the PDB four letter code and optional model numbers and alternate location indicators as part of the URL. For example, PDB code 1ra9 is validated using the URL http://flipper.services.came.sbg.ac.at/cgi-bin/flipper.php?PDBCode=1ra9. A detailed description is provided by the NQ-Flipper online help page.
ACKNOWLEDGEMENTS
We thank Ralf Grosse-Kunstleve for kind permission to use his sglite crystallographic symmetry library. This work was supported by FWF Austria, grant number P13710-MOB. Funding to pay the Open Access publication charges for this article was provided by the University of Salzburg, Austria.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yoder MD, Keen NT, Jurnak F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 2.Battiste JL, Mao HY, Rao NS, Tan RY, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. Alpha helix-RNA major groove recognition in an HIV-1 Rev peptide RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 3.Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 4.Vernet T, Tessier DC, Chatellier J, Plouffe C, Lee TS, Thomas DY, Storer AC, Menard R. Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 1995;270:16645–16652. doi: 10.1074/jbc.270.28.16645. [DOI] [PubMed] [Google Scholar]
- 5.Raymond AC, Rideout MC, Staker B, Hjerrild K, Burgin AB. Analysis of human tyrosyl-DNA phosphodiesterase I catalytic residues. J. Mol. Biol. 2004;338:895–906. doi: 10.1016/j.jmb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 7.Evdokimov AG, Anderson DE, Routzahn KM, Waugh DS. Structural basis for oligosaccharide recognition by Pyrococcus furiosus maltodextrin-binding protein. J. Mol. Biol. 2001;305:891–904. doi: 10.1006/jmbi.2000.4202. [DOI] [PubMed] [Google Scholar]
- 8.Schafer K, Magnusson U, Scheffel F, Schiefner A, Sandgren MO, Diederichs K, Welte W, Hulsmann A, Schneider E. X-ray structures of the maltose-maltodextrin-binding protein of the thermoacidophilic bacterium alicyclobacillus acidocaldarius provide insight into acid stability of proteins. J. Mol. Biol. 2004;335:261–274. doi: 10.1016/j.jmb.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 9.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weichenberger CX, Sippl MJ. Self-consistent assignment of asparagine and glutamine amide rotamers in protein crystal structures. Structure. 2006;14:967–972. doi: 10.1016/j.str.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Weichenberger CX, Sippl MJ. NQ-Flipper: validation and correction of asparagine/glutamine amide rotamers in protein crystal structures. Bioinformatics. 2006;22:1397–1398. doi: 10.1093/bioinformatics/btl128. [DOI] [PubMed] [Google Scholar]
- 12.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 13.McDonald IK, Thornton JM. The application of hydrogen bonding analysis in X-ray crystallography to help orientate asparagine, glutamine and histidine side chains. Protein Eng. 1995;8:217–224. doi: 10.1093/protein/8.3.217. [DOI] [PubMed] [Google Scholar]
- 14.Hooft RW, Sander C, Vriend G. Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins. 1996;26:363–376. doi: 10.1002/(SICI)1097-0134(199612)26:4<363::AID-PROT1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 15.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 16.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sippl MJ. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J. Mol. Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 18.Chandler D. Introduction to Modern Statistical Mechanics. New York: Oxford University Press; 1987. [Google Scholar]
- 19.Sippl MJ. Helmholtz free energy of peptide hydrogen bonds in proteins. J. Mol. Biol. 1996;260:644–648. doi: 10.1006/jmbi.1996.0427. [DOI] [PubMed] [Google Scholar]
- 20.Sippl MJ, Ortner M, Jaritz M, Lackner P, Flockner H. Helmholtz free energies of atom pair interactions in proteins. Fold Des. 1996;1:289–298. doi: 10.1016/S1359-0278(96)00042-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Huang Q, Teng M, Weeks CM, Jelsch C, Zhang R, Niu L. The crystal structure of a novel, inactive, lysine 49 PLA2 from Agkistrodon acutus venom: an ultrahigh resolution, ab initio structure determination. J. Biol. Chem. 2003;278:41400–41408. doi: 10.1074/jbc.M305210200. [DOI] [PubMed] [Google Scholar]