Abstract
ProSight PTM 2.0 (http://prosightptm2.scs.uiuc.edu) is the next generation of the ProSight PTM web-based system for the identification and characterization of proteins using top down tandem mass spectrometry. It introduces an entirely new data-driven interface, integrated Sequence Gazer for protein characterization, support for fixed modifications, terminal modifications and improved support for multiple precursor ions (multiplexing). Furthermore, it supports data import and export for local analysis and collaboration.
INTRODUCTION
In recent years, top down tandem mass spectrometry (MS/MS) has become more popular as a technique for the identification and characterization of proteins. A common approach to top down MS/MS relies on high-mass resolution measurements of the protein precursor ions and the resulting fragment ions. These measurements allow the inference of neutral mass data from the m/z spectra. For large precursor ions, DECON can be used to determine the precursor mass (1,2) while fragment ions are often processed with either THRASH (3), or a manufacturer-specific algorithm like Xtract (4). ProSight PTM is a tool for using the neutral mass data inferred from mass spectra to identify and characterize proteins.
ProSight PTM 1.0 (5) was developed as a web-based application that enabled researchers using neutral mass lists of precursor and fragment ions to query annotated proteomic databases (6,7). When combined with known or predicted PTM information, this would allow them to identify and characterize proteins by determining which known proteins could both have the observed precursor mass and result in the observed fragment pattern. Two types of database schema were supported: a simple schema and a highly annotated schema. Simple schema databases took into account only sequence variants and a few special phoshphorylation (8) cases. Highly annotated schema databases, on the other hand, took into account a large number of potential post-translational modifications (9,10), alone and in combination with others (6,7). By querying the observed neutral masses against these databases, a user could accomplish protein identification and characterization using the top down approach.
We report the deployment of version 2.0 of ProSight PTM. It is based around a fundamentally new model of user interaction, significantly improving usability and data organization. New features also include tools for rapid manual protein characterization, improvements in database annotation, support for additional fixed modifications, and a unified XML-based file format for importing and exporting data.
IMPLEMENTATION
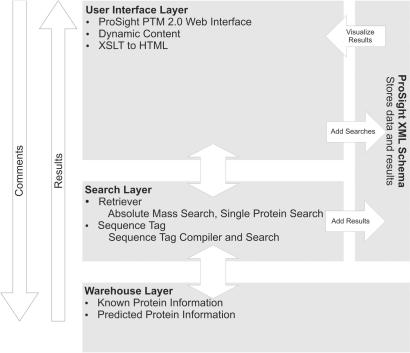
The system is built as a three-layer stack, consisting of independent web-based user interface, search and data warehouse layers (Figure 1). During operation, user searches are converted into XML descriptions, which are pushed to the search layer, consisting of database search engines such as Retriever (5). The search layer then constructs the appropriate queries, issues them to the data warehouse layer, consisting of a set of MySQL database schemas, and integrates the results into the XML-based search descriptions. These are then pushed back up to the user interface layer, where the web interface is generated dynamically based on XSLT (XSL transformations) of the underlying XML.
Figure 1.
The System Architecture of ProSight PTM 2.0.
The user interface, and indeed the whole system, is organized around the concept of an MS/MS experiment. An experiment is a set of neutral mass observations of at least one precursor ion and at least one fragment ion. Associated with this data are one or more searches. Each search is the property of an experiment; each search result is the property of a search. The user experience is thus of analyzing individual experiments by adding searches, and then interpreting their results. This is a marked contrast to the previous version, where the dominant usage paradigm was the processing of data files to produce new data files.
The interface reflects the underlying XML-based data structure of ProSight PTM 2.0. The user may import or export experiments in ProSight Upload Format (PUF), an XML schema designed for representing top down experiments. PUF files may be imported into neuroProSight (http://neuroprosight.scs.uiuc.edu) for neuropeptide searching or ProSightPC (Thermo Fisher, San Jose) for local standalone searching, searched, and the results exported back to ProSight PTM 2.0. Because they are XML-based, PUF files are entirely human-readable with no additional tools beyond a text editor.
Tools and analyses are now fully integrated into the data display. Rather than loading a tool and selecting the specific data file for it to operate on, the user now simply selects a particular experiment or result; the data is displayed with all possible actions the user may take with that data. This makes the interface highly intuitive, and enables the user to focus on the data, rather than the tools. There are, however, a number of new tools and functionalities that merit specific note.
Scalable fragment maps
All search hits are now presented as scalable fragment maps, which may be copied into any document. Because there is no longer any need to generate large raster files, even the largest hit lists have fragment maps generated for all hits, with no negative impact on bandwidth utilization. Scalable fragment maps are constructed as a text block with custom glyphs. These glyphs are stored as a TrueType (11) font file which users must download and install prior to using the system. As TrueType fonts are designed to be scalable, the glyphs may be drawn to any size without loss of quality.
Sequence Gazer
ProSight PTM 2.0 supports integrated manual protein characterization functionality via Sequence Gazer (12). This interactive tool enables the user to quantitatively test hypotheses about the PTM profile of a protein against intact and fragment ion mass data. Any search hit may be loaded into Sequence Gazer with a single click, characterized and the best characterization saved as a completed Single Protein search.
Annotation expansion
The databases underlying ProSight PTM 2.0 have been standardized on the ‘highly-annotated’ schema (5). Shotgun Annotation is used to populate the highly annotated schema with potential protein forms based on the base sequence, potential sequence variability and potential PTMs. Due to this improvement, all searches now support PTMs. Additionally, a significant improvement has been made to the human proteome database. By integrating SNP data from dbSNP (13,14), over 30 000 unique coding SNPs have been added into the database. This considerably improves scores for those proteins containing cSNPs from primary human cells or tissues.
Fixed modifications
A fixed modification is one that is found on all instances of a given residue in a protein. They are typically applied during sample preparation and alter a protein's observed and fragment ion masses. Fixed modifications may be applied to Absolute Mass or Single Protein searches (Table 1). The set of supported modifications is readily extensible, and users are encouraged to send desired fixed modifications to prosightptm@scs.uiuc.edu so that they may be added to the supported set.
Table 1.
Currently supported fixed modifications
| Cysteine fixed modifications | Acrylamide cysteine |
| BME cysteine | |
| Iodoacetamide cysteine | |
| Vinylpyridine cysteine | |
| Methionine fixed modifications | Sulfone methionine |
| Sulfoxide methionine |
Terminal modifications
Terminal modifications are those that are found exclusively at the termini of a protein. These may be separated into two classes: common and user-selected. Common modifications are N-terminal acetylation, N-terminal formylation and initial methionine cleavage. The protein forms arising from common modification combinations are always included in the database, regardless of whether or not any outside source describes these forms. Likewise, they are always included in searches. User-selected terminal modifications, on the other hand, are only included in the database if they were noted in an outside source. They may be included in a search at user discretion. C-terminal amidation is currently available as a user-selected terminal modification.
OPERATION
Prior to logging in for the first time, the user must first download the ProSight font from the ProSight PTM 2.0 home page. This will allow fragment maps to be properly displayed. Once this is done, the user is free to create new experiments and analyze data.
Creating an experiment
As described previously, all data and analyses in the system are organized into experiments. The user may create an experiment either by manually entering the data and parameters, or by importing an already defined experiment. If they choose to manually enter the data, the user will select Add Experiment from the Tools menu. Upon doing so, the Experiment Adder page will be loaded, and the user will be prompted to define the experiment. To fully define an experiment, the user must specify three sets of parameters: experiment data, intact ion data and fragment ion data. Experiment data is limited to the fragmentation method used during data acquisition. Different fragmentation methods result in different types of ions being generated. The system supports three types of fragmentation methods: collision-induced dissociation (CID), infrared multiphoton dissociation (IRMPD) and electron capture dissociation (ECD) (15). Intact-ion data consists of the neutral masses of one or more precursor ions and their mass type parameter. The system supports both monoisotopic and average masses. These may be entered directly, or in the form of a MIDAS peak list (PKL) file (16). Fragment-ion data likewise consists of the neutral masses of one or more fragment ions, their mass type parameter and intensities. Again, these may be entered directly, or as a PKL file. It should be noted that PKL files contain additional data beyond what the system requires: monoisotopic m/z and average m/z. Although unused, this data is stored and propagated, and the user may retrieve it at a later date. If entered manually, unused data is initialized to default values (typically ‘1’). The user may also choose to enter a descriptive experimental comment.
An alternative method for adding a new experiment is to import a ProSight Upload Format (PUF) file. To do so, the user must select Import Experiments from the Tools menu, select the appropriate file, and click ‘Import’. Multiple experiments may be added in one pass using the import dialog, since a PUF file can contain multiple MS/MS experiments. This allows the user to share experiments between ProSight PTM 2.0, neuroProSight and ProSightPC (Thermo Fisher, San Jose).
Modifying experiments
Once an experiment has been created, it is possible to edit its data. The four editable components are: the list of precursor masses, the list of fragment masses, the user comments and noise reduction. Each of these actions may be taken by selecting the appropriate icon on the toolbar for the experiment that is to be added (Figure 2). While the comment may be edited in-place, the other functions load a separate tool. The Mass Editor is loaded in the case of manual mass list modification, and Noise Reducer is loaded in the case of noise reduction. The Mass Editor allows the editing of each attribute of the mass list, including unused attributes. The Noise Reducer has been described previously (5).
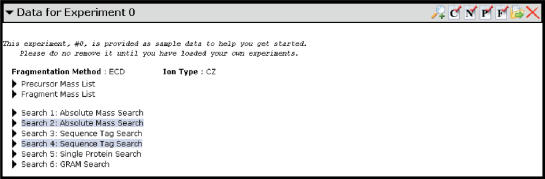
Figure 2.
A typical experiment entry. An experiment is displayed in a pane with a title bar. On the right side of the title bar is the toolbar with seven icons. From left to right, the icons are: Add Experiment, Edit Comment, Reduce Noise, Edit Precursor Mass, Edit Fragment Mass, Export Experiment and Delete Experiment. Clicking on the experiment title expands the experiment details. In general, selecting any list or search will expand it. For instance, selecting the ‘Precursor Mass List’ will expand it into a table of all precursor ion masses.
Adding searches
An arbitrary number of searches may be associated with a particular experiment. A search can either be added based on the raw data, or on the results of another search. Three types of searches may be added: Absolute Mass, Sequence Tag (17) and Single Protein (5). In order to create one of these searches, one must navigate to the experiment desired, and select the Add Search icon on that experiment's toolbar. This will load the Search Adder (12). Once a search is added to an experiment, it appears under that experiment's search list, pending and collapsed into a brief descriptive entry. Pending search entries are highlighted blue, while finished searches are not highlighted. Upon selecting an entry (Figure 3), it expands into a description of the type of search, search parameters, comments and a number of controls representing operations that may be done on the search. For all searches, the user may edit the comments, remove the search and generate a printable report. For pending searches, context-sensitive controls appear below the search parameters that represent actions that may be done on that type of search. Absolute Mass and Single Protein searches have a single control for running the search. Sequence Tag searches have a number of possible operations, depending on the context. If a Sequence Tag search was created indicating use of the ProSight PTM sequence tag compiler, one may either compile the sequence tags or compile and run the search. Once the tags are constructed (either after compilation or if they were entered manually), a regular Run Search control appears. In any case, selecting Run Search takes the user to an automatically refreshing page which will display the status of the search. Once the search is done, a link is displayed to take the user back to the data manager. If any hits have been found, they will appear as a collapsed list beneath the search parameters for that search. If multiple precursor ions were included in the data for that experiment, separate hit lists will be created for each precursor.
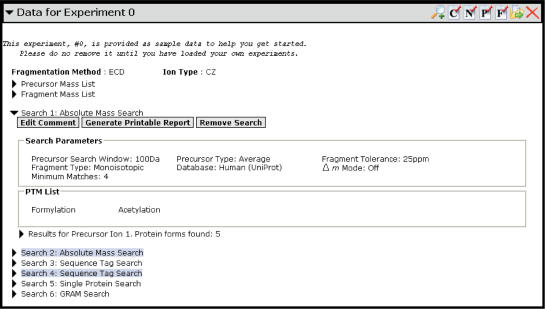
Figure 3.
A typical completed search entry. The PTM list describes the PTMs that were chosen for searching when this search was constructed. Below the PTM list is the list of results for this search. If this had been a pending search, instead of a list of results, a Run Search control would have been displayed instead.
Results display and data-driven search creation
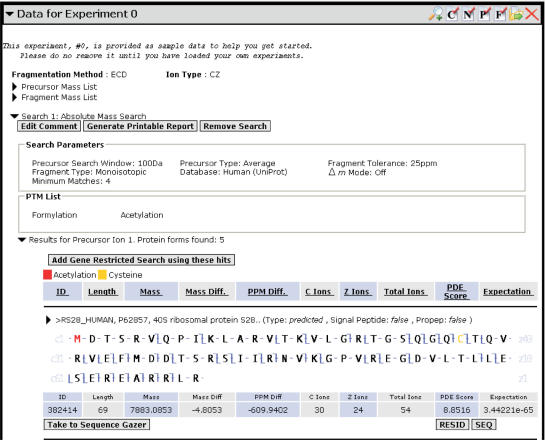
Once the search is run, all hits are automatically stored as a collapsible list for each precursor ion, under the relevant search entry (Figure 4). All search hits are displayed as a scalable fragment map. Above the fragment map is the description of the search hit and below the map are a set of search-dependent hit metrics. A matching fragments table is generated for a hit by selecting the toggle adjacent to the description. The hit metrics for Absolute Mass and Single Protein search hits include hit mass, mass difference with the observed precursor in Daltons and p.p.m., number of matching ions (B or C, Y or Z and total), PDE score (18) and expectation value (19). For Sequence Tag searches, hit metrics are the protein form ID and score. All search hits may have their raw sequence (SEQ) or PTM-marked sequence in RESID (20) form (RESID) displayed.
Figure 4.
Hit Details. A typical result, consisting of a fragment map, a data table and a set of controls.
A number of further actions may be taken with search hits. All search results have a ‘Take to Sequence Gazer’ control below them. This loads that particular search hit into Sequence Gazer, a manual protein characterization tool. After characterization is complete in Sequence Gazer, the saved results appear as a Single Protein search under the relevant experiment.
Absolute Mass and Sequence Tag search hit lists have an additional control that appears above them: ‘Add Gene Restricted Search using these hits’. This loads the search adder to create a Gene Restricted Absolute Mass search using that result list. This appears in the search list as a ‘GRAM Search’. GRAM search result lists may be operated on as any other Absolute Mass search results. The GRAM search was previously known as a ‘Hybrid’ search in ProSight PTM 1.0.
Finishing analysis
Once analysis is complete, search results may be exported in two ways. First, the experiment as a whole may be exported as a PUF file for later review in either ProSight PTM 2.0 or ProSightPC. Second, a printable report may be generated for the particular search of interest. This report takes the form of a webpage that may be printed, saved or imaged for other use.
To export a PUF file, the user selects the ‘Export Experiment’ icon on the toolbar of the experiment to be exported. They will be immediately prompted to save the resulting PUF file. This file may be saved for evaluation or backup, uploaded to neuroProSight for searching or loaded into ProSightPC for local searching. The PUF file contains all searches (run or pending), all search hits and all data that was originally used to create the experiment.
OBTAINING AN ACCOUNT
Accounts for academic users are provided free of charge. Email prosightptm@scs.uiuc.edu for account information.
FUTURE DIRECTIONS
We intend to continue adding highly annotated databases to ProSight PTM 2.0, increasing the number of species whose proteomes may be searched. Specific species and annotations may be requested by users. Furthermore, currently under development are databases that optimally present the proteomes of higher Eukarya in silico via Shotgun Annotation. This will improve both search speed and the characterizing power of the average top down search.
ACKNOWLEDGEMENTS
This work has been supported by National Institutes of Health NIDA grant P30 DA 018310-039002 and GM 067193. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health NIDA grant P30 DA 018310-039002.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fenn JB. Ion formation from charged droplets: roles of geometry, energy, and time. J. Am. Soc. Mass Spectromet. 1993;4:524–535. doi: 10.1016/1044-0305(93)85014-O. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JR, Meng F, Forbes AJ, Cargile BJ, Kelleher NL. Fourier-transform mass spectrometry for automated fragmentation and identification of 5–20 kDa proteins in mixtures. Electrophoresis. 2002;23:3217–3223. doi: 10.1002/1522-2683(200209)23:18<3217::AID-ELPS3217>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 3.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J. Am. Mass Spectromet. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 4.Zabrouskov V, Senko MW, Du Y, Leduc RD, Kelleher NL. New and automated MSn approaches for top-down identification of modified proteins. J. Am. Soc. Mass Spectromet. 2005;16:2027–2038. doi: 10.1016/j.jasms.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeDuc RD, Taylor GK, Kim Y-B, Januszyk TE, Bynum LH, Sola JV, Garavelli JS, Kelleher NL. ProSight PTM: an integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Res. 2004;32:W340–W345. doi: 10.1093/nar/gkh447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth MJ, Forbes AJ, Boyne MT, II, Kim Y-B, Robinson DE, Kelleher NL. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol. Cell. Proteom. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc. 2004;126:3386–3387. doi: 10.1021/ja039748i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 9.Washburn MP, Yates J.R., III Analysis of the microbial proteome. Curr. opinion Microbiol. 2000;3:292–297. doi: 10.1016/s1369-5274(00)00092-8. [DOI] [PubMed] [Google Scholar]
- 10.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 11.Herman D, et al. CA: Cupertino: Apple Computer, Inc; 1996. The TrueType Reference Manual. [Google Scholar]
- 12.LeDuc RD, Kelleher NL. Using ProSight PTM and related tools for targeted protein identification and characterization with high mass accuracy tandem MS Data. Current Protocols in Bioinformatics. 2007. Unit 13.6. [DOI] [PubMed]
- 13.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 14.Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 16.Senko MW, Canterbury JD, Guan S, Marshall AG. A high-performance modular data system for Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectromet. 1996;10:1839–1844. doi: 10.1002/(SICI)1097-0231(199611)10:14<1839::AID-RCM718>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 17.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 18.Reid GE, Shang H, Hogan JM, Lee GU, McLuckey SA. Gas-phase concentration, purification, and identification of whole proteins from complex mixtures. J. Am. Chem. Soc. 2002;124:7353–7362. doi: 10.1021/ja025966k. [DOI] [PubMed] [Google Scholar]
- 19.Meng F, Cargile BJ, Miller LM, Forbes AJ, Johnson JR, Kelleher NL. Informatics and multiplexing of intact protein identification in bacteria and the archaea. Nat. Biotechnol. 2001;19:952–957. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
- 20.Garavelli JS, Hou Z, Pattabiraman N, Stephens RM. The RESID Database of protein structure modifications and the NRL-3D Sequence-Structure Database. Nucleic Acids Res. 2001;29:199–201. doi: 10.1093/nar/29.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]