Abstract
Asterias (http://www.asterias.info) is an open-source, web-based, suite for the analysis of gene expression and aCGH data. Asterias implements validated statistical methods, and most of the applications use parallel computing, which permits taking advantage of multicore CPUs and computing clusters. Access to, and further analysis of, additional biological information and annotations (PubMed references, Gene Ontology terms, KEGG and Reactome pathways) are available either for individual genes (from clickable links in tables and figures) or sets of genes. These applications cover from array normalization to imputation and preprocessing, differential gene expression analysis, class and survival prediction and aCGH analysis. The source code is available, allowing for extention and reuse of the software. The links and analysis of additional functional information, parallelization of computation and open-source availability of the code make Asterias a unique suite that can exploit features specific to web-based environments.
INTRODUCTION
Web-based applications are well suited for the analysis of microarray and genomic data. They do not require the user to install or upgrade any software, the computational capabilities (a concern with the large data sets common in genomic studies) are not limited by the user's hardware (only by the server) and, with the recent advances in web technologies, can offer a user interface and experience very similar to that of desktop applications. Integrated suites that carry out a complete set of analyses of several different types of data can be very appealing for many users, as the applications within the suite present a similar interface, have homogeneous input requirements and allow the analysis of various types of data that many wet-lab researchers deal with routinely (e.g., from microarray data normalization to aCGH). In addition, web-based tools offer the opportunity to quickly bring new methodological developments to many potential users. Therefore, there is room for additional work in integrated web-based suites to incorporate key statistical and methodological advances.
Web-based tools: requirements and desirable features
Web-based tools do not need to compromise on statistical rigor and can use validated and state-of-the-art methods. When trying to discover differentially expressed genes, multiple testing problems should be taken into account (1,2) and, since many microarray studies are really observational studies with human patients, it is often necessary to include additional clinical covariates to minimize confounding problems (3,4). In addition, we can also borrow information from all genes in the array when carrying out the test for each gene, using moderated statistics and Empirical Bayes approaches (5). When dealing with classification and prediction, it is crucial to avoid biases that lead to overoptimistic estimates of the error rates. These biases include ‘selection bias’ (6,7) and bias caused by selecting and reporting the error rate of the classifier (among a set of classifiers) with the smallest cross-validated error rate (8,9). Additionally, gene selection in the context of classification often yields many solutions with similar prediction errors, but which share few common genes (10–12); being unaware of the possible instability of our results can lead to a false sense of certainty that the given set is special and distinct.
In addition to statistical rigor, a modern tool should incorporate the increasing availability of multicore processors and clusters built with off-the-shelf components, which are probably the major opportunities for significant performance gains in the near future (13,14). MPI (15) is one approach to parallelize computations over several CPUs and/or processor cores, thus decreasing execution time. Interestingly, web-based applications are well suited for this task; if deployed in a computing cluster, the parallelization, while transparent for the user, permits harvesting computational resources that are rarely available to individual researchers.
To help in the interpretation of results (16,17), web-based tools are ideally suited to link to additional sources of information, such as PubMed references, gene ontology (GO) terms, and the UCSC and Ensembl databases and KEGG and Reactome pathways. Moreover, it is possible to carry out further analysis with this additional information, such as highlighting features (e.g. pathways, GO terms, etc.) that might be characteristic of a set of selected genes that are, say, very common among the genes that tend to be repeatedly selected as relevant for a classification problem. This usage of additional information can help us understand whether there are biological commonalities behind the possible multiple solutions (see above).
Finally, the availability of source code, under an open-source license, allows other researchers to further improve the method and provide bug fixes, use the code for instruction and teaching, permits to verify claims by method developers, encourages reproducible research, and ensures that the international research community remains the owner of the tools it needs to carry out its work (18). These features facilitate fast methodological development based on previous work, and expedite the transfer of results to applied research. The value of the source code is further enhanced if best practices (19) as well as common open-source practices (including public code repositories and open bug tracking) are followed, ultimately allowing the building of a community of contributors (20).
ASTERIAS: UNIQUE FEATURES
Some of the currently available web-based suites include RACE (26), MIDAW (27), Gepas (28) and CARMAweb (29). All of these, however, fail one or more of the above requirements. We have thus developed Asterias to fulfill those requirements. First, Asterias is the only web-based application which we know of that is designed, from the beginning, to make extensive use of parallelization in its computations. The speed up can be dramatic when run in a computing cluster (in our own installation of 30 dual-processor servers, some applications speed up by factors of 30 × to 50 ×). Second, Asterias, as with some other suites, includes tools that cover the complete range of needs of many researchers (from normalization to aCGH analysis, including imputation, differential expression and class prediction), but Asterias is the only suite that includes tools for searching for large sets of predictive genes (GeneSrF), and gene selection, molecular signatures and prediction with survival data (SignS). Third, we provide statistically rigorous and state-of-the-art methods, from the well-known BioConductor limma package (5), in the study of differential expression, to the best available methods for aCGH analysis, as reported in recent reviews (30,31). Moreover, we facilitate the analysis of multiple solutions in class prediction and gene selection tools (e.g. frequency of genes in bootstrap and cross-validation runs and similarity of solutions with regards to biological role via an analysis of additional information—see below). Fourth, the development of Asterias includes functional and regression testing of our applications, using publicly available and open-source tests; this is also a unique feature of Asterias.
In addition, the newest release of Asterias includes two important additions. We make (virtually) all of our code available under open-source licenses (GNU GPL and Affero GPL) and have an open-source development mode, including open bug tracking and full repository history available. Finally, an important novelty with respect to our latest release, the user can analyze the results (e.g. the genes that have been selected as good prognosis classifiers) and examine PubMed references, GO terms, KEGG pathways or Reactome pathways for those genes using the new PaLS web server. PaLS, coupled with the examination of multiple solutions, can ease the biological interpretation of the results, specially in studies of gene selection and classification.
Asterias shares some common history with the GEPAS suite (28), and one of the authors of Asterias (RD-U) was heavily involved in the development of GEPAS (32–34) and related tools (35–37). Nowadays, Asterias and GEPAS only share the tool DNMAD—although the R code in Asterias' DNMAD has changed to adapt it to the latest BioConductor releases— and a similar approach to web server load-balancing, via Pound or LVS, with everything else being different. A brief history of the split can be found at http://asterias.bioinfo.cnio.es/Asterias.Gepas.html. The main differences between Asterias and GEPAS are our strong commitment to parallel computing, differences in the type of applications being developed (e.g. SignS, ADaCGH, GeneSrF, PomeloII) and software development mode (all of our code is available under open-source licenses, including complete repositories and functional tests).
FUNCTIONALITY, INPUT, OUTPUT
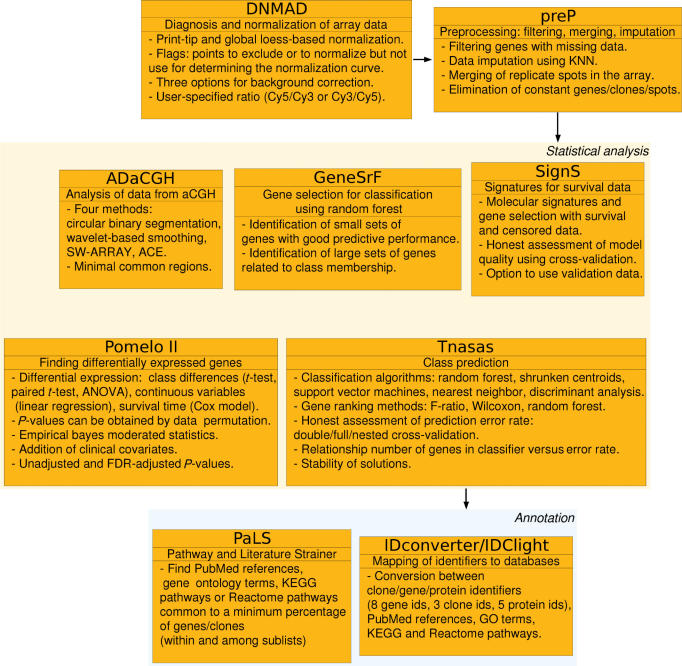
Figures 1 and 2 show the main functionality provided by each of the Asterias applications, the relationships between the tools and the main input and output of each application. All the analysis tools are accessible from preP, but can also be accessed directly, and preP can be accessed either directly or from DNMAD.
Figure 1.
Asterias: functionality and data and information flow between sets of applications (see details in Figure 2). References for ADaCGH methods are: circular binary segmentation (21), wavelet-based smoothing (22), SW-ARRAY (23) and ACE (24). The method implemented in SignS is from (25).
Figure 2.
Asterias: input/output and data and information flow between applications. Black and blue arrows involve files, green arrows URLs. Olive boxes denote graphical output.
Input to all applications are plain text files, with tab-separated columns. Further details are provided in the online help of each application. Output of most applications includes both text-like output, with clickable links to IDClight (38) and PaLS and graphical output. Some applications (e.g. IDconverter) can also provide tabular output in other formats (e.g. Microsoft Excel). Screenshots of output are provided in the Supplementary Data.
IMPLEMENTATION
Most of the statistical functionality is written in R (39), with some code in C/C++ (Pomelo II and several dynamically loadable code in R packages), and extensive use of parallelization using MPI and R interfaces to MPI. The R code uses standard R or BioConductor packages (some of them modified to allow parallel computation) and our own packages (e.g. varSelRF, ADaCGH). Full details on the R and BioConductor packages used are provided in the help pages of each application. The web interfaces and input data validation are written in Python (with some legacy Perl and PHP in DNMAD and IDconverter). Clickable figures and tables are usually generated using R, with additional post-processing using Python. The database server for IDconverter, IDClight and PaLS is MySQL. Scripts for database management and generation are also written in Python. JavaScript is used in several applications, most notably in Pomelo II (AJAX), but also on clickable figures and collapsible trees. Booting and halting the LAM/MPI universes is accomplished by a combination of Python and shell scripts. We create a new LAM/MPI universe for each run of each application, and the actual nodes/CPUs that are used in a LAM/MPI universe are determined at run-time (thus excluding nodes that are down).
Documentation, help, bug tracking
Online help, including tutorials, examples and sample files, is available for all applications. Pomelo II includes additional tutorials as flash movies. The online tutorials and examples are licensed under a Creative Commons license (http://www.creativecommons.org), allowing for redistribution and classroom use. The R packages have, additionally, help available in the standard R format. Bug tracking is available from the Bioinformatics.org project page http://bioinformatics.org/bugs/?group_id=630.
Availability
Our publicly accessible installation runs on a cluster with 30 dual-CPU nodes with Debian GNU/Linux. The web service is load-balanced (we are currently using Linux Virtual Server, but have used Pound in the past), which ensures balancing of the master nodes for MPI and of the non-parallelized applications (e.g. preP). All of the code (except, temporarily, for PaLS) is available under open-source licenses (either GNU GPL v.2 or Affero Public License). The complete repositories can be downloaded from Bioinformatics.org (http://bioinformatics.org/asterias) or Launchpad (https://launchpad.net/asterias). The R package varSelRF is also available from the R repositories.
Testing, maturity and number of accesses
Asterias includes a test suite that uses FunkLoad (http://funkload.nuxeo.org). The test suite tests the user interface, handling of error conditions and incorrectly formated files and the numerical output, and can be run on demand, and wherever new changes are introduced in the software, thus ensuring appropriate quality control and regression testing. The complete code is also available (see ‘Functional testing’ in the repositories). For Pomelo II (which makes extensive use of AJAX), additional tests using Selenium (http://www.openqa.org/selenium/) are available (http://pomelo2.bioinfo.cnio.es/tests.html); these tests verify that the application runs correctly under different operating systems and browsers.
Asterias is a mature suite. Its oldest application, DNMAD (40), has been running since October 2003, and the newest one, PaLS, has been running since October 2006. The rest of the applications have been running for at least a year, often considerably longer. The number of data sets analyzed (note that these are counts of actual numbers of successfully uploaded files, not just hits) in the 10-month period February 1, 2006 and November 30, 2006, range from 3700 and 2900 for preP and Pomelo II, respectively, between to 530 and 340 for SignS and GeneSrF, except for IDconverter and IDClight, which have over 70 daily uses.
Future work
Our main development effort is focused on making Asterias easy to install and deploy, from laptops to clusters of workstations. We are currently re-implementing all of Asterias using Pylons (http://pylonshq.com), a Python web framework, together with installation scripts that ease the configuration, management and monitoring of the computing nodes and parallel computing layers. We are also exploring other languages and paradigms, such as QHTML (41), built on top of Mozart/Oz, to solve the problem that ‘Building web-based applications requires the mastering of a number of languages/technologies (e.g. HTML, CSS, CGI, ASP, PHP, XML, etc.). Such languages and technologies were created to address different aspects on a by-need, evolutionary manner. The result is a plethora of tools that are fitted together in an ad hoc fashion’. (41).
In both cases, our ultimate objective is developing a general framework (or at least a large enough set of case examples) that will make it much simpler for any bioinformatician/biostatistician to take new ideas and developments from the primary methodological research and make them quickly available as web-based applications. These web-based applications should be capable of using advances in computing and hardware (multicore CPUs, computing clusters built with off-the-shelf components, parallel computing and concurrency) and web technologies (e.g., AJAX).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We want to thank the many testers at CNIO and elsewhere for feedback on the applications and for bug reports. Bionformatics.org and Launchpad provided hosting for the repositories. Our applications would not have been possible without the excellent and free R language and its many freely available packages. Funding was provided by Fundación de Investigación Médica Mutua Madrileña and Project TIC2003-09331-C02-02 of the Spanish Ministry of Education and Science (MEC). R.D.-U. is partially supported by the Ramón y Cajal programme of the Spanish MEC. Applications are running on clusters of machines purchased with funds from the RTICCC from the Spanish FIS. Funding to pay the Open Access publication charges for this article was provided by the Fundación de Investigación Médica Mutua Madrileña.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ge Y, Dudoit S, Speed T. Resampling-based multiple testing for microarray data analysis (with discussion) TEST. 2003;12:1–77. [Google Scholar]
- 2.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 3.Potter JD. Epidemiology, cancer genetics and microarrays: making correct inferences, using appropriate designs. Trends Genet. 2003;19:690–695. doi: 10.1016/j.tig.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Díaz-Uriarte R. Supervised methods with genomic data: a review and cautionary view. In. In: Azuaje F, Dopazo J, editors. Data Analysis and Visualization in Genomics and Proteomics. Wiley New York: 2005. pp. 193–214. chapter 12. [Google Scholar]
- 5.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. and Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 6.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl Acad. Sci. USA. 2002;99:6562–6566. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of dna microarray data for diagnostic and prognostic classification. J. Nat. Cancer Inst. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 8.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics. 2006;7(1) doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dudoit S, Fridlyand J. Classification in microarray experiments. In, Statistical Analysis of Gene Expression Microarray Data, chapter 3. In: Speed T, editor. Chapman & Hall New York: 2003. pp. 93–158. [Google Scholar]
- 10.Somorjai RL, Dolenko B, Baumgartner R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics. 2003;19:1484–1491. doi: 10.1093/bioinformatics/btg182. [DOI] [PubMed] [Google Scholar]
- 11.Pan KH, Lih CJ, Cohen SN. Effects of threshold choice on biological conclusions reached during analysis of gene expression by DNA microarrays. Proc. Natl Acad. Sci. USA. 2005;102:8961–8965. doi: 10.1073/pnas.0502674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díaz-Uriarte R, Alvarez deAndrés S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutter H. The free lunch is over: A fundamental turn toward concurrency in software. Dr. Dobb's Journal. 2005;30(3):202–210. [Google Scholar]
- 14.Kontoghiorghes EJ, editor. Handbook of Parallel Computing and Statistics. Boca Raton, FL: Chapman & Hall, CRC Press; 2006. [Google Scholar]
- 15.Pacheco P. Parallel Programming with MPI. San Francisco: Morgan Kufman; 1997. [Google Scholar]
- 16.Hyatt G, Melamed R, Park R, Seguritan R, Laplace C, Poirot L, Zucchelli S, Obst R, Matos M, Venanzi E, et al. Gene expression microarrays: glimpses of the immunological genome. Nat. Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat. Genet. 2005;(37 Suppl):S31–S37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 18.Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- 19.Baxter SM, Day SW, Fetrow JS, Reisinger SJ. Scientific software development is not an oxymoron. PLoS Comput. Biol. 2006;2:e87+. doi: 10.1371/journal.pcbi.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fogel KF. Producing Open Source Software. Sebastopol, CA: O'Reilly; 2005. [Google Scholar]
- 21.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 22.Hsu L, Self SG, Grove D, Randolph T, Wang K, Delrow JJ, Loo L, Porter P. Denoising array-based comparative genomic hybridization data using wavelets. Biostatistics. 2005;6:211–226. doi: 10.1093/biostatistics/kxi004. [DOI] [PubMed] [Google Scholar]
- 23.Price TS, Regan R, Mott R, Hedman A, Honey B, Daniels RJ, Smith L, Greenfield A, Tiganescu A, Buckle V, et al. SW-ARRAY: a dynamic programming solution for the identification of copy-number changes in genomic DNA using array comparative genome hybridization data. Nucleic Acids Res. 2005;33:3455–3464. doi: 10.1093/nar/gki643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingjaerde OC, Baumbusch LO, Liestol K, Glad IK, Borresen-Dale AL. CGH-Explorer: a program for analysis of array-CGH data. Bioinformatics. 2005;21:821–822. doi: 10.1093/bioinformatics/bti113. [DOI] [PubMed] [Google Scholar]
- 25.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, et al. Prediction of Survival in Follicular Lymphoma Based on Molecular Features of Tumor-Infiltrating Immune Cells. N. Engl. J. Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 26.Psarros M, Heber S, Sick M, Thoppae G, Harshman K, Sick B. RACE: Remote Analysis Computation for gene Expression data. Nucleic Acids Res. 2005;33:W638–W643. doi: 10.1093/nar/gki490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romualdi C, Vitulo N, Favero MD, Lanfranchi G. MIDAW: a web tool for statistical analysis of microarray data. Nucleic Acids Res. 2005;33:W644–W649. doi: 10.1093/nar/gki497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montaner D, TÃrraga J, Huerta-Cepas J, Burguet J, Vaquerizas JM, Conde L, Minguez P, Vera J, Mukherjee S, Valls J, et al. Next station in microarray data analysis: Gepas. Nucleic Acids Res. 2006;34:W486–W491. doi: 10.1093/nar/gkl197. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. Carmaweb: comprehensive r- and bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 2006;34:W498–W503. doi: 10.1093/nar/gkl038. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willenbrock H, Fridlyand J. A comparison study: applying segmentation to array cgh data for downstream analyses. Bioinformatics. 2005;21:4084–4091. doi: 10.1093/bioinformatics/bti677. [DOI] [PubMed] [Google Scholar]
- 31.Lai WRR, Johnson MDD, Kucherlapati R, Park PJJ. Comparative analysis of algorithms for identifying amplifications and deletions in array cgh data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrero J, Al-Shahrour F, Díaz-Uriarte R, Mateos Á, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS, a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herrero J, Vaquerizas JM, Al-Shahrour F, Conde L, Mateos Á, Santoyo J, Díaz-Uriarte R, H.Dopazo J. New challenges in gene expression data analysis and the extended GEPAS. Nucleic Acids Res. 2004;32:W485–W491. doi: 10.1093/nar/gkh421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaquerizas JM, Conde L, Yankilevich P, Cabezon A, Minguez P, Diaz-Uriarte R, Al-Shahrour F, Herrero J, Dopazo J. GEPAS, an experiment-oriented pipeline for the analysis of microarray gene expression data. Nucleic Acids Res. 2005;33:W616–W620. doi: 10.1093/nar/gki500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Díaz-Uriarte R, Al-Shahrour F, Dopazo J. The Use of Go Terms to Understand the Biological Significance of Microarray Differential Gene Expression Datain Methods of Microarray Data Analysis III, papers from Camda '02, Kluwer; 2003. pp. 233–247. [Google Scholar]
- 36.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 37.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. Discovering molecular functions significantly related to phenotypes by combining gene expression data and biological information. Bioinformatics. 2005;21(13):2988–2993. doi: 10.1093/bioinformatics/bti457. [DOI] [PubMed] [Google Scholar]
- 38.Alibés A, Yankilevich P, Cañada A, Diaz-Uriarte R. Idconverter and idclight: conversion and annotation of gene and protein ids. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing Vienna, Austria (2004)
- 40.Vaquerizas JM, Dopazo J, Díaz-Uriarte R. DNMAD: web-based diagnosis and normalization for microarray data. Bioinformatics. 2004;20:3656–3658. doi: 10.1093/bioinformatics/bth401. [DOI] [PubMed] [Google Scholar]
- 41.El-Ansary S, Grolaux D, Van Roy P, Rafea M. Van Roy P. Overcoming the multiplicity of languages and technologies for web-based development using a multi-paradigm approach. In. Multiparadigm Programming in Mozart/OZ. 2005:113–124. chapter 10, Springer. [Google Scholar]