Abstract
SNPs located within the open reading frame of a gene that result in an alteration in the amino acid sequence of the encoded protein [nonsynonymous SNPs (nsSNPs)] might directly or indirectly affect functionality of the protein, alone or in the interactions in a multi-protein complex, by increasing/decreasing the activity of the metabolic pathway. Understanding the functional consequences of such changes and drawing conclusions about the molecular basis of diseases, involves integrating information from multiple heterogeneous sources including sequence, structure data and pathway relations between proteins. The data from NCBI's SNP database (dbSNP), gene and protein databases from Entrez, protein structures from the PDB and pathway information from KEGG have all been cross referenced into the StSNP web server, in an effort to provide combined integrated, reports about nsSNPs. StSNP provides ‘on the fly’ comparative modeling of nsSNPs with links to metabolic pathway information, along with real-time visual comparative analysis of the modeled structures using the Friend software application. The use of metabolic pathways in StSNP allows a researcher to examine possible disease-related pathways associated with a particular nsSNP(s), and link the diseases with the current available molecular structure data. The server is publicly available at http://glinka.bio.neu.edu/StSNP/.
INTRODUCTION
SNPs represent one of the most common forms of genetic variation in a population (1,2). Currently, (December 2006) the public SNP database (dbSNP) (3) contains 11.9 million SNP candidates, of which 5.6 million have been validated. Nonsynonymous SNPs (nsSNPs), the SNPs located within the open reading frame of a gene that result in an alteration in the amino acid sequence of the encoded protein might directly or indirectly affect protein functionality alone or its interactions in a multi-protein complex, by increasing/decreasing the activity of the metabolic pathway (1,4). nsSNPs have been linked to a wide variety of diseases; affecting protein function, altering DNA and transcription factor binding sites, reducing protein solubility and destabilizing protein structures (4). Therefore, understanding the functional consequences of nonsynonymous changes and predicting potential causes and the molecular basis of diseases involves integration of information from multiple heterogeneous sources including sequence, structure data and pathway relations between proteins.
SNP information is currently collected in several databases, including: dbSNP, the Human Genome Variation Database (HGVbase) (5), the Japanese Single Nucleotide Polymorphism (JSNP) database (6) and the HapMap Project (1). Currently, there is a number of studies and resources which have begun to explore the effects of nsSNPs on the tertiary structure of proteins and their functionality, including: SNPs3D (7), PolyPhen (8), TopoSNP (9), ModSNP (10), LS-SNP (11), SNPeffect (12), MutDB (13,14) and Snap (15), have all been released for public use. We have provided a brief description of the available resources for SNP analysis in Tables 1 and 2. It should be noted, this is not a comparison table but a reference table, as the field is in its infancy and all resources are currently evolving, with each database having strengths.
Table 1.
Representing query and modeling options for resources
| SNPs3D | PolyPhen | topoSNP | SNPeffect | LS-SNP | MutDB | SNP@Domain | Snap | StSNP | |
|---|---|---|---|---|---|---|---|---|---|
| Keyword search is available | Yes | No | No | Yes | No | Yes | No | Yes | Yes |
| Modeling is available | Yes (precomputed) | No | Yes (precomputed) | No | Yes (precomputed) | No, mutations are highlighted in the structure | No, highlighting the amino acids affected by SNPs. | No | Yes (on the fly) |
| Pathway Information | Yes | No | No | No | Yes | No | No | Gene relations are available | Yes |
| Search by Protein ID | Yes | Yes | No | Yes | Yes | Yes | No | Yes | Yes |
| Search by FASTA sequence | No | Yes | Yes | No | No | No | No | No | No |
| Search by PDB ID | Yes | No | No | Yes | No | No | No | No | Yes |
| Analysis of nsSNPs | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | No |
| Connection with OMIM or nsSNP info | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | No |
| Graphic display of the nsSNPs on protein sequence | No | No | No | No | No | Yes | Yes | Yes | Yes |
| Search by pathways | No | No | No | No | Yes | No | No | No | Yes |
| Disease Related info available | Yes | Yes | Yes | Yes | Yes | No | Yes(link to OMIM) | Yes | No |
| Domain info for the proteins | No | Yes (links available) | No | Yes (links available) | Yes (link to SCOP) | No | Yes | Yes | No |
| Search by rs# | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes |
| Search by gene | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes |
Table 2.
Table shows the differences and the similarities of the resources for their search options and background information
| SNPs3D | PolyPhen | TopoSNP | SNPeffect | LS-SNP | MutDB | SNP@Domain | Snap | StSNP | |
|---|---|---|---|---|---|---|---|---|---|
| Description | Website which assigns molecular functional effects of nsSNPs based on structure and sequence analysis. | An automatic tool for prediction of possible impact of an amino acid substitution on the structure and function of a human protein. | Provides an online resource for analyzing nsSNPs that can be mapped onto known 3D structures of proteins. These include disease associated nsSNPs derived from OMIM db and other nsSNPs derived from dbSNP. | an online resource of human ns SNPs (nsSNPs) mapping phenotypic effects of allelic variation in human genes. | Maps nsSNPs onto protein sequences, functional pathways and comparative protein structure models and predicts positions where nsSNPs destabilize proteins, interfere with the formation of domain-domain interfaces, have an effect on protein-ligand binding, or severely impact human health. | MutDB is to annotate human variation data with protein structural information and other functionally relevant information. | SNP@Domain is a web resource, to identify SNPs within human protein domains. | A medical-focus database which can provide user comprehensive information of a single gene and relationship between genes based on SNPs. | Compare structural nsSNP distributions in many proteins or protein complexes. StSNP enables to researchers to map nsSNPs onto protein structures and visualize their structural locations by using the multiple structure-sequence viewer Friend. StSNP includes human nsSNPs. |
| Sources | dbSNP, HGMD, PDB, PQS, OMIM, SwissPro, LocusLink, GO, KEGG, Mouse Knockout | HGVbase,PDB, dbSNP | dbSNP, OMIM, PFAM | dbSNP, NCBI-Protein, PDB, GO, Pfam, OMIM, CSA, SwitchPDB | dbSNP, SwissProt/TrEMBL, LocusLink, PDB | Swiss-Prot and dbSNP, PDB | SCOP, Pfam, Ensembl database, dbSNP | EnsEMBL, UCSC, Swiss-Prot, Pfam, DAS-CBS, KEGG, MINT, BIND, OMIM | dbSNP, KEGG, PDB, HapMap, LocusLink, NCBI-Protein |
| Links | HGMD, dbSNP, OMIM, PubMed, KEGG | almost all the possible links after the selection of a structure | PDB | Several links (almost all the possible links). | SwissProt/TrEMBL, USCSGenome_browser for SNPs, PDBsum, SCOP | dbSNP, NCBI-Protein, -mrna,-nucleotide, Swiss-Prot | Ensemble, SCOP, Pfam, SIFT, OMIM | EnsEMBL, OMIM | dbSNP, PDB, NCBI Protein, NCBI Chromosome Map |
| Display Info | Gene info, SNP info from HGDV, visualization by Jmol/RasMol Applet | Info for the query only entered SNP location and substitutions, Prediction (benign or damaging), scoring | nsSNP info, available PDB info, Visualization of mapping of nsSNPs is available with MDL's Chime Plugin | nsSNP info, rs#, functional sites, cellular processing of the protein, PDB structure, disease relation, structure information of the protein with wild type and changed aa. | SwissPro ID, rs#, nsSNP info, interface domain info. | Graphic display of exons, SNPs and related data. | Graphic display of domain and SNPs, 2D and 3D visualization, links to external sources. | Gene relation view, disease list, polymorphism statistics, gene sequence, primer parameters, domain info, sequence primer design, gene info | Graphic display of alignment, Protein and nsSNP info, rs#, pathway info, graphic display of pathways. Visualization of modeling is available with Friend software. |
| Visualization | Viewable with Jmol | Not available | Viewable in the Chime window. It will need MDL's Chime Plugin to view. | Not available | RasMol | PyMOL, UCSF's Chimera | MDL Chime Plugin | Gene relation view by GraphViz | Visualization of modeling is available with Friend viewer. |
| User Friendliness | Easy to use, visualization of the pages easy, everything can be seen all at once in a summary | Difficult and results are coming in too many pages | Easy to use, query page is straight forward. | Easy to use, everything can be seen in a summary, detail information is available. | Easy to use and very explicit | Easy to use, results page is compact enough to reach data conveniently | Clear and well summarized results page | Well summarized but it still takes time to navigate. | Easy to use, Steps are easy to identify and understand. (biased by AU) |
| Update Status | Not available | No info | Regular intervals (no specific timing) depending on the sources | Regulary updated (no specific info) | Once or two times a year | Regular intervals, depending on the sources | - (no info) | Updated on a regular basis following the updates on the major sources, dbSNP, PDB, KEGG and others. | |
| How many nsSNPs exist in db? | 50 772, 29 485 can be modeled. | 50 919, 44 005 (unique rs entries) | 27 417 | 23 426 proteins with SNPs | 28 000 validated SNP | 1487 SNPs annotated with protein structures (based on related article) | 17 639 SNPs within SCOP and 28 238 SNPs within Pfam domains were identified. | 68 072 | 33 692 |
| Query options | rs#, gene name, protein accession id, keyword | Fasta input with position and substitution info, PDB,PQS,sorting by E-value or identity, threshold contact. | Fasta format of protein sequence, options disease and nondisease associated nsSNPs. | rs#, RefSeq, SwissProt/TrEMBL, PDB, EC#, OMIM#, text search Filters with PDB or Disease Related | SwissPro, rs#, KEGG, HUGO gene, chromosome range | Keyword, gene symbol, refseq protein-id, rna-id, Swiss-Pro Gene Name | rs#, gene name/symbol, domain name/id | Keywords, rs#, protein,accession code from Ensembl and Uniprot/SWISSPROT, chromosome areas, Markers or clones | Protein Accesion id, PDB ID, keyword, rs#, metabolic pathways,gene, length, substitution search |
| Modeling | In house comparative modeling program (based on Modeller's approach) | Not applicable | Not mentioned | Not applicable | MODELLER | The mutations are highlighted in the structure. | Highlighting the amino acids affected by SNPs. | Not applicable | MODELLER |
| Cross References | AVAILABLE | AVAILABLE | AVAILABLE | AVAILABLE | AVAILABLE | AVAILABLE | AVAILABLE | AVAILABLE | AVAILABLE |
| Validated nsSNPs only | BOTH | BOTH | BOTH | OPTIONAL | BOTH | BOTH | BOTH | BOTH | BOTH |
| How many nsSNPs available to map by one search? | all available nsSNP can be mapped. | Not applicable | User dependable | Not applicable | 1 nsSNP per model | The mutations are highlighted in the structure. | Highlighting the amino acids affected by SNPs. | Not applicable | All available nsSNPs can be mapped. |
We present StSNP, a web-based server, which provides the ability to analyze and compare human nsSNP(s) in protein structures, protein complexes and protein–protein interfaces, where nsSNP and structure data on protein complexes are available in PDB, along with the analysis of the metabolic data within a given pathway. Usually nsSNP do not inactivate protein functionality completely, otherwise the mutation would most likely be lethal, instead nsSNPs change the protein activity at some level, either directly (occurring close to active site) or indirectly through interactions with other proteins in the pathway; therefore, such information has to be considered mutually. As a result, we have developed StSNP, which utilizes information from different sources and provides ‘on the fly’ comparative modeling of the wild-type and mutated proteins (when an appropriate structural template is available) along with real-time analysis and visualization of structures and sequences (16) to assist researchers in visual inspection of the possible effects of the nsSNPs in protein structure. StSNP enables users to analyze data in different formats by utilizing different search capabilities, by keyword, NCBI protein accession numbers, PDB IDs (17) and NCBI nsSNP ids quickly retrieve targeted information.
DESIGN AND IMPLEMENTATION SOURCES
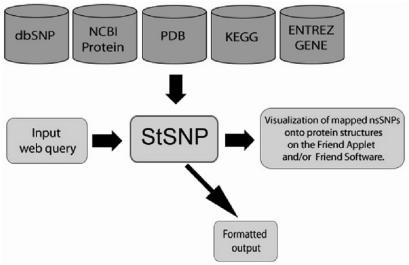
In general, the internal database structure has been inherited from the Structural Exon database (SEDB) (18). StSNP was implemented using a MySQL database running on a Linux server, with PERL scripts used for all data retrieval and output (Figure 1). StSNP utilizes three major data sources: (1) Protein sequences from NCBI, (2) the reference and nsSNPs locations from NCBI's dbSNP and (3) structures and sequences from the PDB. Every protein sequence has a pre-calculated list of structural modeling templates found by BLAST (19), and stored in a database for quick retrieval. The actual aligning of the protein sequence and the PDB sequence was implemented with the Smith–Waterman algorithm (20,21), using similarity specific scoring matrices, from BLOSUM30 to BLOSUM90 (22). The pathway information is utilized from KEGG (23,24), human gene/protein information is gathered from NCBI's Entrez Gene (25), and the comparative modeling phase is done by MODELLER (26). The modeling part of StSNP is interactive and allows the user to choose a template from the list, select particular mutations to be modeled, calculate the model and subsequently visualize the superimposition of the models and template in the Friend applet. Additionally, simultaneous analysis of structurally similar proteins/models for structural correlation of nsSNP locations can be done in the Friend applet by the TOPOFIT structure alignment method (27,28). StSNP currently contains 33 692 nsSNPs, 14 858 protein sequences, 12 741 genes and 25 617 protein structures.
Figure 1.
StSNP is an interactive web server, which utilizes several heterogeneous data sources.
WEB SERVER FEATURES
StSNP has several types of search options, including search by a Protein ID, PDB ID or keyword, all of which together integrates nsSNP related information. For example, the Protein ID search displays the known nsSNP(s) for the protein, while the PDB ID search provides a list of similar Protein IDs with nsSNP(s). Both searches will provide a link to pathway information if the data is available. The resulting report pages provide the user with options for model template selection. Only templates satisfying the following two criteria are shown: the nsSNP(s) has to be within the alignment of the protein sequence with template and the sequence identity of the alignment has to be ≥30%. The modeling step provides the user with the ability to choose which nsSNPs to map, and after completion, a user can instantly visualize the models with the Friend applet. StSNP has several browsing and search capabilities as well, for example, searching for available structures by protein length and percent similarity, or by a specifically chosen reference and nonsynonymous residue within a particular chromosome. The features found in StSNP have been design with graphics, plots and easily readable tables with the end user in mind.
EXAMPLES OF USE
Mapping nsSNPs on to protein structures
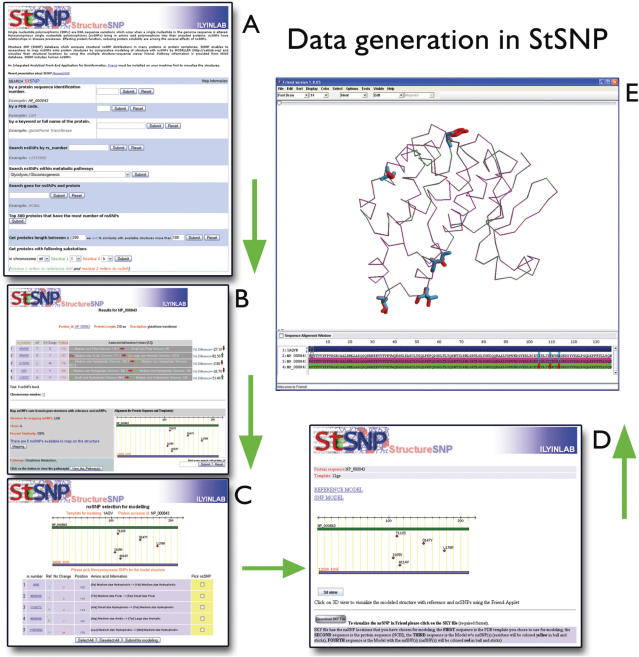
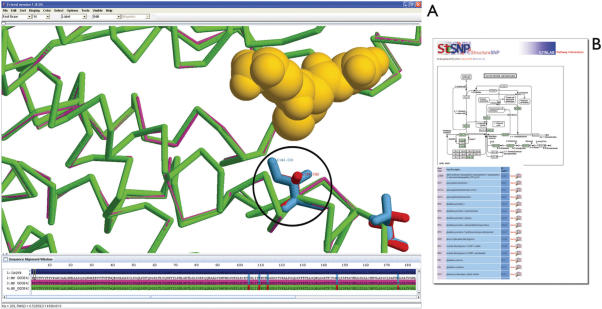
Results shown in Figure 2 were generated with the query Glutathione S Transferase (GST, Protein ID NP_000843), a family of multifunctional enzymes involved in cellular detoxification of xenobiotics and reactive endogenous compounds of oxidative metabolism (29). The output page reports the available reference and nonsynonymous residues for the protein with the rs number, amino acid properties for the variations, and the alignment picture of protein sequence with template including nsSNP locations. In this example, all nsSNPs are located inside the alignment and thus available for mapping onto PDB ID 1aqv chain B. The next step is to choose the nsSNPs for modeling. All the known nsSNPs associated with GST, I105V, T110S, A114V, D147Y and L176M have been modeled in this example and are presented in Figure 3A. A black circle denotes where isoleucine has changed to valine at position 105. The role of functional I105V GSTP1 polymorphism in the pathogenesis of methamphetamine abuse was studied, with researchers noting that individuals with the G allele (valine) are expected to have decreased GST detoxification (29). It is visible from the mapping of this nsSNP onto the protein structure (Figure 3A) the location of I105V is located in direct contact with the glutathione, and could potentially have a strong effect on the GST activity or its binding affinity with glutathione. The results section also provides a user with a link to glutathione metabolism in order to view other members found in the pathway (Figure 3B).
Figure 2.
Data generation in StSNP. (A) Main query page, (B) Formatted data for nsSNPs along with graphical alignment representation, (C) nsSNP(s) selection for modeling, (D) Output page, and (E) Visualization in the Friend applet.
Figure 3.
(A) Glutathione S Transferase is shown with nsSNP locations displayed in ball and stick representation, with I105V marked with a black circle. The reference residues are shown in blue, nonsynonymous residues in red and the substrate glutathione is displayed in space fill representation (yellow). The query for the example was Protein ID NP_000843 and template PDB ID 1aqv chain B. (B) The Results section also provides a user with a link to glutathione metabolism in order to view other members found in the pathway.
Another example, Aldehyde Dehydrogenase-2 (ALDH2) (PROTEIN ID NUMBER= NP_000681) is illustrated in Figure 4. ALDH2 is involved in acetaldehyde oxidation at physiological concentrations and found when a person consumes alcohol. Worldwide, the Lys504 allele has the highest prevalence (30–50%) in Asian populations (30). In this example, glutamate is replaced by lysine at position 504 (Glu504Lys), where it has been demonstrated to essentially eliminate ALDH2 activity (31). From these examples, one can see how a quick search in StSNP in conjunction with the structural mapping of the nsSNP locations provides structural support to the medical studies mentioned here and may facilitate in the designing of future experiments.
Figure 4.
Aldehyde dehydrogenase-2 is shown with nsSNP locations displayed in ball and stick representation, with E504K marked with a black circle. The reference residues are shown in blue, nonsynonymous residues in red and the substrate NAD is displayed in space fill representation (green). The query for the example was Protein ID NP_000681 and template PDB ID 1ag8 chain A.
CONCLUSIONS
StSNP provides practical, user friendly access to the wealth of information related to nsSNPs by seamlessly connecting various databases into one pipeline. Key functional and structural information along with known pathways the proteins are involved in, have all been linked together to provide users some advantages when compared to other current resources: (a) the sequence, structure and pathway information have all been cross-referenced, which enables a user to quickly query and visualize the inter-related nsSNP data; (b) a graphical display of the nsSNPs provides a user with the location of the nsSNP(s) in terms of primary sequence, and whether such nsSNP(s) can be modeled; (c) the modeling options provide the user with a choice of which nsSNP to map and visualize which nsSNPs could potentially have deleterious effects on a protein's function; (d) the modeled protein structures are automatically loaded in Friend, where they can be easily viewed, compared and analyzed; (e) finally, StSNP will be updated on a regular basis following the updates on the major sources, dbSNP, PDB, KEGG and others.
Thus, the first steps have been taken in the development of a resource for mapping nsSNPs onto protein structures, providing structural insight into the effects of nsSNPs on proteins such as, stability, functionality, protein–protein interactions and other structurally related issues. As a web server in a rapidly evolving area of research, StSNP is designed to evolve with other related resources; future directions include; a more detailed analysis of the SNP, predictions of the functional/biological implications of the SNP(s) and the use of image map technology from the KEGG API for more interactive data retrieval. StSNP creates the basis for further studies involving the metabolic pathways and the disease(s) associated with a particular SNP.
ACKNOWLEDGEMENT
The Open Access publication charges for this manuscript were waived by Oxford University Press. Funding to pay the Open Access charges for this paper were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 2.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 3.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 4.Chasman D, Adams RM. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: structure-based assessment of amino acid variation. J. Mol. Biol. 2001;307:683–706. doi: 10.1006/jmbi.2001.4510. [DOI] [PubMed] [Google Scholar]
- 5.Fredman D, Siegfried M, Yuan YP, Bork P, Lehvaslaiho H, Brookes AJ. HGVbase: a human sequence variation database emphasizing data quality and a broad spectrum of data sources. Nucleic Acids Res. 2002;30:387–391. doi: 10.1093/nar/30.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirakawa M, Tanaka T, Hashimoto Y, Kuroda M, Takagi T, Nakamura Y. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 2002;30:158–162. doi: 10.1093/nar/30.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Moult J. SNPs, protein structure, and disease. Hum. Mutat. 2001;17:263–270. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- 8.Sunyaev S, Ramensky V, Koch I, Lathe W, III, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 9.Stitziel NO, Binkowski TA, Tseng YY, Kasif S, Liang J. topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res. 2004;32:D520–D522. doi: 10.1093/nar/gkh104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yip YL, Scheib H, Diemand AV, Gattiker A, Famiglietti LM, Gasteiger E, Bairoch A. The Swiss-Prot variant page and the ModSNP database: a resource for sequence and structure information on human protein variants. Hum. Mutat. 2004;23:464–470. doi: 10.1002/humu.20021. [DOI] [PubMed] [Google Scholar]
- 11.Karchin R, Diekhans M, Kelly L, Thomas DJ, Pieper U, Eswar N, Haussler D, Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;12:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 12.Reumers J, Schymkowitz J, Ferkinghoff-Borg J, Stricher F, Serrano L, Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33:D527–D532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dantzer J, Moad C, Heiland R, Mooney S. MutDB services: interactive structural analysis of mutation data. Nucleic Acids Res. 2005;33:W311–W314. doi: 10.1093/nar/gki404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han A, Kang HJ, Cho Y, Lee S, Kim YJ, Gong S. SNP@Domain: a web resource of single nucleotide polymorphisms (SNPs) within protein domain structures and sequences. Nucleic Acids Res. 2006;34:W642–W644. doi: 10.1093/nar/gkl323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Ma L, Li H, Vang S, Hu Y, Bolund L, Wang J. Snap: an integrated SNP annotation platform. Nucleic Acids Res. 2007;35:D707–D710. doi: 10.1093/nar/gkl969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abyzov A, Errami M, Leslin CM, Ilyin VA. Friend, an integrated analytical front-end application for bioinformatics. Bioinformatics. 2005;21:3677–3678. doi: 10.1093/bioinformatics/bti602. [DOI] [PubMed] [Google Scholar]
- 17.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, et al. The Protein Data Bank. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 18.Leslin CM, Abyzov A, Ilyin VA. Structural exon database, SEDB, mapping exon boundaries on multiple protein structures. Bioinformatics. 2004;20:1801–1803. doi: 10.1093/bioinformatics/bth150. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Smith TF, Waterman MS. Comparison of biosequences. Adv. Appl. Math. 2005;2:482–489. [Google Scholar]
- 21.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 22.Henikoff S, Henikoff JG. Performance evaluation of amino acid substitution matrices. Proteins. 1993;17:49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa M. A database for post-genome analysis. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M. The KEGG database. Novartis. Found. Symp. 2002;247:91–101. [PubMed] [Google Scholar]
- 25.Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 27.Ilyin VA, Abyzov A, Leslin CM. Structural alignment of proteins by a novel TOPOFIT method, as a superimposition of common volumes at a topomax point. Protein Sci. 2004;13:1865–1874. doi: 10.1110/ps.04672604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslin CM, Abyzov A, Ilyin VA. TOPOFIT-DB, a database of protein structural alignments based on the TOPOFIT method. Nucleic Acids Res. 2007;35:D317–D321. doi: 10.1093/nar/gkl809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto T, Hashimoto K, Matsuzawa D, Shimizu E, Sekine Y, Inada T, Ozaki N, Iwata N, Harano M, Komiyama T, et al. A functional glutathione S-transferase P1 gene polymorphism is associated with methamphetamine-induced psychosis in Japanese population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;135:5–9. doi: 10.1002/ajmg.b.30164. [DOI] [PubMed] [Google Scholar]
- 30.Goedde HW, Agarwal DP, Harada S, Meier-Tackmann D, Ruofu D, Bienzle U, Kroeger A, Hussein L. Population genetic studies on aldehyde dehydrogenase isozyme deficiency and alcohol sensitivity. Am. J. Hum. Genet. 1983;35:769–772. [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Zhang D, Jin W, Shao C, Yan P, Xu C, Sheng H, Liu Y, Yu J, et al. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J. Clin. Invest. 2006;116:506–511. doi: 10.1172/JCI26564. [DOI] [PMC free article] [PubMed] [Google Scholar]