Abstract
Here we present Primer3Plus, a new web interface to the popular Primer3 primer design program as an enhanced alternative for the CGI- scripts that come with Primer3. Primer3 consists of a command line program and a web interface. The web interface is one large form showing all of the possible options. This makes the interface powerful, but at the same time confusing for occasional users. Primer3Plus provides an intuitive user interface using present-day web technologies and has been developed in close collaboration with molecular biologists and technicians regularly designing primers. It focuses on the task at hand, and hides detailed settings from the user until these are needed. We also added functionality to automate specific tasks like designing primers for cloning or step-wise sequencing. Settings and designed primer sequences can be stored locally for later use. Primer3Plus supports a range of common sequence formats, such as FASTA. Finally, primers selected by Primer3Plus can be sent to an order form, allowing tight integration into laboratory ordering systems. Moreover, the open architecture of Primer3Plus allows easy expansion or integration of external software packages. The Primer3Plus Perl source code is available under GPL license from SourceForge. Primer3Plus is available at http://www.bioinformatics.nl/primer3plus.
INTRODUCTION
Oligonucleotide primers are widely used in various molecular biology techniques like DNA sequencing and the polymerase chain reaction (PCR). Since a primer serves as the starting point for DNA replication, specific binding of the oligonucleotide to the target sequence on the template strand is essential for a successful experiment. The binding specificity of a primer is determined by several of its properties, like the melting temperature (Tm), GC-content and self complementarity. Designing primers is usually done with the help of computer programs, among which Primer3 is most widely used judging from the hundreds of citations of the primary publication (1). Primer3 is popular since the program can be used online and is redistributed free of charge. Recently, a SourceForge project was started for Primer3 in which several improvements are being discussed and implemented. Subsequently Primer3 was updated to include an additional method for Tm calculation [SantaLucia model, (2)], since it was argued that primer design based on the Breslauer model shows variation in the predicted Tm (3,4).
Primer3 consists of a command line C program, and an HTML web interface written in Perl. The web interface is one large form showing all possible options. This makes it powerful, but at the same time confusing for the occasional user; without in-depth knowledge it is hard to tell which of the settings are important for a specific design task. Some tasks, like designing PCR primers for position specific cloning, are not easy to perform with the current web interface, since this would require multiple runs of the program under different settings. Because of these issues with the current web interface and facilitated by its liberal license, several academic and commercial parties have created their own command line and/or web interface to Primer3, tailored to their specific needs and audience; e.g. EMBOSS’ EPRIMER3 and the PCR Suite (5,6). We took this effort one step further and developed a general purpose, easy to use and powerful new web interface for Primer3, called Primer3Plus that is available at http://www.bioinformatics.nl/primer3plus.
WEB INTERFACE
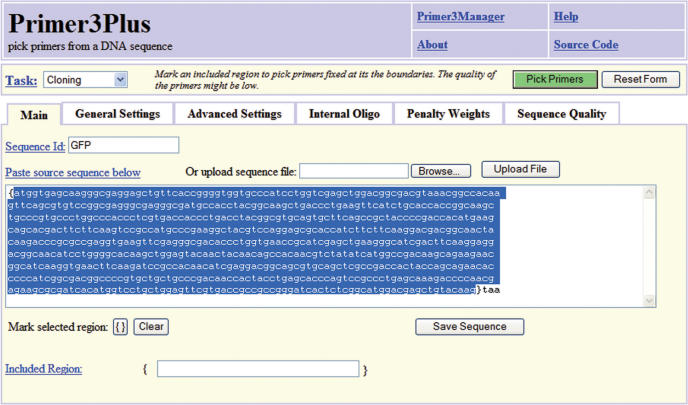
We aimed to design the Primer3Plus web interface in such a way that it is comprehensible for occasional users, and yet powerful for the experienced users that need to do more complex or laborious tasks. Where the original Primer3 web interface presents all options and settings in one large web page, Primer3Plus starts with a more simple screen containing only the input boxes for the sequence and the option to select the target region for amplification (Figure 1). The target sequence can be pasted or uploaded in any common format, such as FASTA and EMBL. Furthermore, the ‘Pick Primers’ button is moved to the top of the page to make sure it is always available. Also a new task selection box is included (top left corner), that provides the possibility to select between five different scenarios for which Primer3Plus can be used (see subsequently).
Figure 1.
Screenshot of the Primer3Plus start page. In contrast to the original Primer3 web interface, detailed settings that are not frequently used are hidden behind tabbed panels. A pull-down task menu is added (top left) that provides five scenarios for which Primer3Plus can be used: gene detection, primers for cloning purposes, sequencing, list of all possible primers and check option for already available primers. Here an example sequence (GFP) was loaded, the Cloning task selected and the open reading frame marked as included region.
For most purposes the default parameter settings are adequate. Adjusting these parameters is possible by using the tabbed panels. The tabs are ordered in the most likely frequency of use, with the left most panel being the start page, followed by the general settings, advanced settings, internal oligo options, penalty weights and sequence quality. The parameter names were kept the same as in the original Primer3 interface to minimize time needed to get familiar with using Primer3Plus. The labels of the parameters are web links to a context-sensitive help text, just like in the original Primer3 web interface. Primer3Plus adds to this a floating tool tip with a brief description for some of the most prominent parameters. Next to the default settings, the user can choose specific settings for special tasks like primer design for qPCR applications. It is also possible to store custom settings locally to save the user from having to go through the configuration each time primers are designed for specific non-standard conditions, like AT-rich organisms or high-salt environments.
The Primer3Plus results page shows the suggested primers with their characteristics, ordered from best to worst. The best scoring primer pair is marked on the template sequence. The selected primers can be submitted to the Primer3Manager where the user can manage a primer collection. Designed primers are stored on the web server for a limited period, allowing users to design multiple primer pairs and combine them in a single order. Users, or actually web clients, are identified using cookies that expire after a set period (currently one week). Primers can also be saved locally as a FASTA file and previously saved primers can be uploaded allowing the addition of new primers to an existing primer collection. Within the Primer3Manager selected primers can be re-analyzed, submitted to the NCBI BLAST service (7) or combined on an order form. In case Primer3Plus is run on a local server, the order form can be customized allowing tight integration into a laboratory primer order management system.
DESIGN TASKS
We discriminate five distinct primer design user scenarios, or tasks, that have been integrated in the new interface. These tasks can be selected using a drop down list at the top of the Primer3Plus page. Currently the tasks include Detection, Cloning, Sequencing, Primer Check and Primer List. More tasks might be added in future versions of the Primer3Plus web interface.
Detection
The Detection task can be used for designing standard PCR primers or hybridization oligos to detect a given sequence. The user provides the template sequence and optionally indicates target regions within the sequence or changes the parameters for primer picking. By pressing the Pick Primers button a list of suggested primer pairs is presented, ordered with the best pair at the top. The Detection task is most reminiscent to the original Primer3 web interface.
Cloning
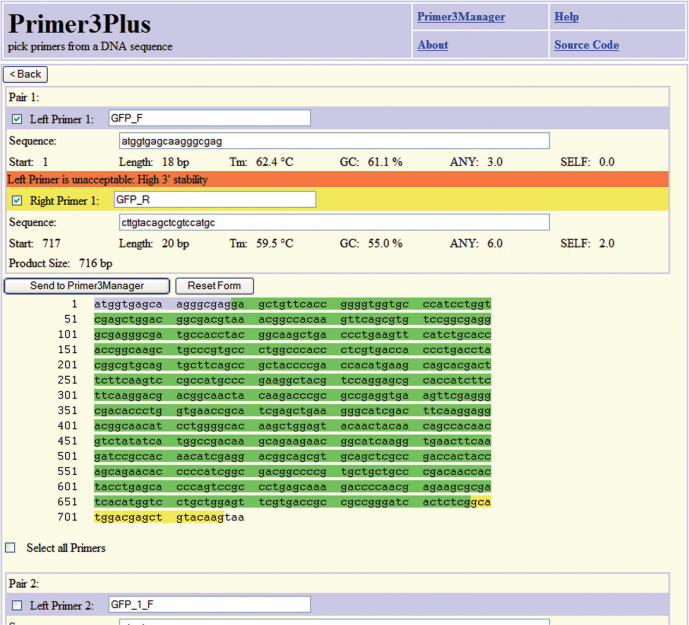
For cloning a PCR product in a specific reading frame in an expression vector, it is important to control precisely the start and end of the PCR product. With the Cloning task either the 5′ or the 3′ ends of the primers can be fixed to the boundary of the included region, marked with curly braces. The output lists the predicted best fitting primer pairs that start or end at the given position. Subsequently, the user can modify the primer sequence, e.g. by adding a restriction enzyme recognition site that facilitates cloning of the PCR product. Fixing primer ends significantly limits the number of possible candidates that fulfil the stringent default settings. If this is the case, alternative primers are given that are accompanied by a warning concerning the relaxed stringencies that have been applied (Figure 2).
Figure 2.
Result page of a Primer3Plus Cloning run showing the left and right primers in blue and yellow. The included region is colored green. Although the left primer is of low quality due to high 3′ stability, it is the best primer starting at this position with the given settings. Alternative primer pairs of lower quality are shown below the sequence (truncated here).
Sequencing
The Sequencing task is developed to design a series of primers on both the forward and reverse strands that can be used for custom primer-based (re-)sequencing of clones. Under default settings, the primers are spaced by a 500 nt interval on both strands. The spacing of primers is configurable within the Advanced settings tab, as is the overlap between the forward and reverse primers. A configurable number of nucleotides immediately following the primer that usually cannot be read reliably, are taken into account when selecting the forward and reverse primers.
Primer Check
The Primer Check task can be used to obtain information on a specified primer, like its melting temperature or self complementarity. This can, for instance, be useful when certain primers are to be reused under different experimental conditions or if the original primer characteristics were lost. The Primer Check option is also available on the Primer3Manager page, enabling a re-check of primers that have been modified by the user, e.g. for cloning purposes. When this task is selected, no template sequence is required and only the primer sequence has to be provided.
Primer List
With the Primer List task, all possible primers that can be designed on the target sequence and meet the current settings will be returned with their corresponding characteristics. This task basically allows manual selection of primers, which could be of interest in case of specific tasks that are not (yet) implemented in Primer3Plus (like picking overlapping primers). The run time of the Primer List task can be relatively long when compared to the other Task options, and can take up to a minute, especially when lengthy target sequences are submitted.
TECHNOLOGY
The Primer3Plus web pages consist of HTML with JavaScript for interactivity. The JavaScript is not essential for proper function of Primer3Plus and can be disabled. Custom settings are stored locally using the Boulder IO format (a simple data format: tag = value). The Primer3Plus web interface has been tested to run properly on the most recent versions of Firefox, Internet Explorer, Safari and Konqueror. The server side software has been written in Perl, using the CGI standard for communicating with the Apache web server. It runs on both SuSE Enterprise Linux 9 and Windows XP, and should run unaltered on any modern Unix/Linux platform.
Primer3Plus is designed with interoperability in mind. It is relatively easy to integrate with other (web) applications. Settings, like the template sequence, can be passed in via HTML POST or GET operations, so applications can be linked directly to an already filled Primer3Plus form.
FUTURE WORK
An annotated view of the template DNA sequence showing, for instance, the open reading frames or intron/exon boundaries would facilitate finding the regions of interest for amplification or detection. For this, Primer3Plus will need to be extended with functionality to extract and display features from uploaded sequence files.
Primer3Plus was recently included in the Primer3 SourceForge project (http://sourceforge.net/projects/primer3/). This allows for tight integration of the web interface with the core program and creates a platform for both developers and end users to discuss further modifications and additions to Primer3Plus.
ACKNOWLEDGEMENTS
The authors would like to thank Steve Rozen for helpful suggestions and ongoing support of Primer3, and Catarina Cardoso and Gerben Bijl for extensive beta testing. R.G. was funded by NWO-VIDI (864.06.007) and NWO-HORIZON (050-71-052). H.N. was supported by the BioAssist program of the Netherlands Bioinformatics Centre. Funding to pay the Open Access publication charges for this article was provided by Wageningen University and Research Centre.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ, USA: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 2.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm091. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 3.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon PMK, Sensen CW. Osprey: a comprehensive tool employing novel methods for the design of oligonucleotides for DNA sequencing and microarrays. Nucleic Acids Res. 2004;32:e133. doi: 10.1093/nar/gnh127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice P, Longden I, Bleasby A. EMBOSS: the european molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 6.Van Baren MJ, Heutink P. The PCR suite. Bioinformatics. 2004;20:591–593. doi: 10.1093/bioinformatics/btg473. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden, T.L. Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]