Abstract
RADAR is a web server that provides a multitude of functionality for RNA data analysis and research. It can align structure-annotated RNA sequences so that both sequence and structure information are taken into consideration during the alignment process. This server is capable of performing pairwise structure alignment, multiple structure alignment, database search and clustering. In addition, RADAR provides two salient features: (i) constrained alignment of RNA secondary structures, and (ii) prediction of the consensus structure for a set of RNA sequences. RADAR will be able to assist scientists in performing many important RNA mining operations, including the understanding of the functionality of RNA sequences, the detection of RNA structural motifs and the clustering of RNA molecules, among others. The web server together with a software package for download is freely accessible at http://datalab.njit.edu/biodata/rna/RSmatch/server.htm and http://www.ccrnp.ncifcrf.gov/~bshapiro/
INTRODUCTION
RNA molecules play various roles in the cell (1–4). Their functionality depends not only on the sequence information but to a large extent on their secondary structures. It would be more cost effective if one were able to determine RNA structure by computational means rather than by using biochemical methods. So, the development of computational predictive approaches of RNA structure is essential (5–8). RNA structure prediction is usually based on the thermodynamics of RNA folding (4,6,9–11) or phylogenetic conservation of base-paired regions (7,8,12–15).
Here, we present a web server, RADAR (acronym for RNA Data Analysis and Research), which performs a multitude of functions related to RNA structure comparison, including pairwise structure alignment, constrained structure alignment, multiple structure alignment, database search, clustering and consensus structure prediction. Our aim behind developing this web server is to develop a versatile tool that provides a computationally efficient platform for performing several tasks related to RNA structure. RADAR has been developed using Perl-CGI and Java. In each run, the server can accept at most 50 RNA sequences or secondary structures for pairwise structure alignment and constrained structure alignment and at most 10 RNA sequences or secondary structures for the other functions where each sequence or structure has at most 300 bases, though the downloadable version does not have this restriction. For the sample data provided by the server, it takes a few seconds for most of the server's functions to complete and display results on the web. It takes about one minute to produce a multiple structure alignment when RNA sequences are fed as input. The database search function needs several minutes to search the Rfam database (http://www.sanger.ac.uk/Software/Rfam/); the results of this function are returned to the user via email, rather than on the web.
METHOD
RADAR employs the RSmatch algorithm (16) for computing the alignment of two RNA secondary structures. Briefly, it decomposes each RNA secondary structure into a set of basic structure components that are further organized by a tree model. With this model, pseudoknots are not allowed. A dynamic programming algorithm is employed to align the two RNA secondary structures. RSmatch is capable of performing both global and local alignment of two RNA secondary structures. The time complexity of the algorithm is O(mn), where m and n are the sizes of the two structures, respectively. This method is an efficient solution to the problem of RNA structure alignment.
By using this structure comparison algorithm, we developed different functionalities such as pairwise structure alignment, multiple structure alignment, database search, clustering, constrained structure alignment and consensus structure prediction, and incorporated these functionalities into RADAR. Pairwise structure alignment involves the alignment of a query structure with each of the subject structures in a set. Multiple structure alignment uses the same alignment algorithm along with a position specific scoring matrix to build up an alignment by including one structure at a time until no appropriate structure can be included in the alignment (16). Database search is done by aligning a query structure one by one with the consensus structures of the non-coding RNA families stored in the release 8.0 of Rfam (17) to find the consensus structures similar to the query structure. This function returns the top k hits as the search result, where k is an adjustable parameter. Clustering is done to compute and display a similarity matrix for a set of RNA secondary structures.
We also developed a constrained version of RNA structure alignment to improve the sensitivity of the alignment. This allows the user to annotate a region of an input RNA structure to be conserved. The conserved region, or constraint, is incorporated into the alignment process to produce biologically more meaningful alignment results. We also implemented a new method to compute the consensus structure for a group of closely related RNA sequences. Details of the two methods are explained below.
Constrained structure alignment
This method constructs the alignment between a query structure and a set of subject structures based upon the knowledge of conserved regions in the query structure. The alignment score is dynamically varied so as to utilize the information of the conserved regions. The alignment computed this way is able to detect structural similarity more accurately. The method comprises two main parts:
(i) Annotating a region in the query RNA structure as conserved
Each position of the conserved region in the query RNA structure is marked using a special character ‘*’ underneath the position. This is termed binary conservation since any position in the query RNA structure is treated to be either 100% conserved (if it is marked with ‘*’) or not conserved at all. If it is found, from wet lab experiments or other sources, that a particular RNA structure contains a motif that we want to search for in other RNA structures in a data set, then that particular RNA structure can be used as a query structure and that motif region can be marked to be conserved in the query structure. Under this circumstance, the user adopts binary 0/1 conservation. Another technique of applying constraints to structure alignment is using the concept of sequence logos (18). With this technique, the degree of conservation at each position of the query RNA structure is a value between 0% and 100%. Details of this technique, which is not implemented in the web server due to its complicated input format but is included in the downloadable version, are given in Supplementary Material.
(ii) Utilization of information about the conserved region
Two cases occur as we compute the alignment score between a query structure and a set of subject structures where the query structure contains marked conserved regions.
Comparison between non-conserved regions: In this case the score assigned is the regular score that is derived from the scoring matrix used by RSmatch.
Comparison involving conserved regions: Here, we multiply the score obtained from the scoring matrix used by RSmatch by a factor λ that will cause the score to either increase or decrease by the λ value. This factor λ is determined by the type of conservation as discussed in more detail in the subsection on ‘Scoring scheme’.
Scoring scheme
The factor by which the score should get magnified or diminished to take into account the conserved region is determined based upon the following: (i) the length of the conserved region; (ii) the length of the whole RNA sequence; (iii) the type of conservation that has been indicated and (iv) any special conditions/preferences decided by the user.
In the default scenario, where knowledge about conservation is not used, the score is directly taken from the scoring matrix employed in RSmatch. For the binary conservation case, the default value for the factor λ is λ = 2 – L/N where L is the length of the conserved region and N is the length of the whole RNA sequence. This ratio is then subtracted from a constant value (2, arbitrarily chosen) so that the bonus/penalty is inversely proportional to the length of the conserved region. If the conservation based on sequence logos is used, it is spread over 0–100%, as described earlier, and these percentage values are passed along with the query RNA structure to the scoring engine and the alignment score varies based on these values.
Consensus structure prediction
This method works in four steps, as described below (experimental results are included in Supplementary Material).
Determine individual RNA structures: For the input RNA sequences, compute their structures having energies that fall within a particular range of the minimum energy using the Vienna RNA package's RNAsubopt function (9). Therefore, for each sequence there can be more than one possible structure. The result consists of the predicted RNA structures for all the RNA sequences in the input file.
Compute a pairwise scoring matrix: In this step, compute the pairwise alignment scores between all structures except for the structures that represent the same RNA sequence. The result is a matrix that gives the score of alignment for every pair of structures. The score of comparison between RNA structures of the same sequence is entered as 0.
Select one structure for each RNA sequence: From the matrix produced in step 2, select the pair of structures that have the best score. These structures are then the chosen structures for the RNA sequences they correspond to. The pairwise scoring matrix is modified to eliminate all the other structures of these RNA sequences. Once again the same process of selecting the best pair of structures and then eliminating the other structures of the sequences they belong to is carried out. This is repeated until we are able to select a structure for each of the input sequences.
Predict the common RNA substructure: This step deals with predicting the consensus RNA substructure that is common to as many RNA sequences in the input file as possible. This is obtained by computing a multiple structure alignment of the RNA structures selected in step 3.
WEB SERVER
The RADAR web server together with a standalone downloadable version is freely available at http://datalab.njit.edu/biodata/rna/RSmatch/server.htm and http://www.ccrnp.ncifcrf.gov/~bshapiro/. A comprehensive help manual is available on this website as well. This online document provides detailed instructions explaining the use of the web server.
Input
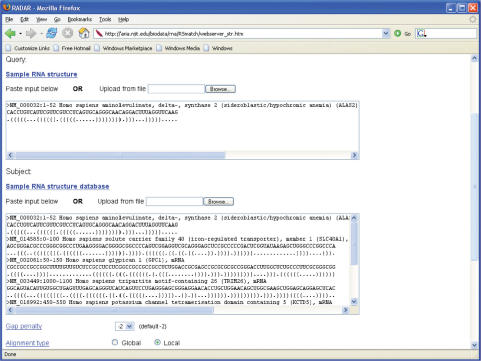
RADAR accepts, as input data, either RNA sequences in the standard FASTA format or RNA secondary structures in the Vienna style Dot Bracket format (9). The input data can be stored in a file to be uploaded to the server or entered directly into the text boxes provided by the server. Figure 1 shows the input interface of RADAR for aligning an RNA secondary structure with a set of subject structures. When RNA sequences are fed as input, RADAR invokes Vienna RNA v1.4 (9) to fold the sequences into RNA secondary structures. Based upon the function chosen, there are different alignment parameters such as gap penalty, scoring matrix, alignment type (global or local) or folding parameters such as minimum free energy, sliding window size, etc. that can be customized by the user. For performing constrained structure alignment, we require the user to annotate the query RNA structure to indicate which region is conserved by marking the region with ‘*’.
Figure 1.
The input interface of RADAR for aligning an RNA secondary structure with a set of subject structures.
Output
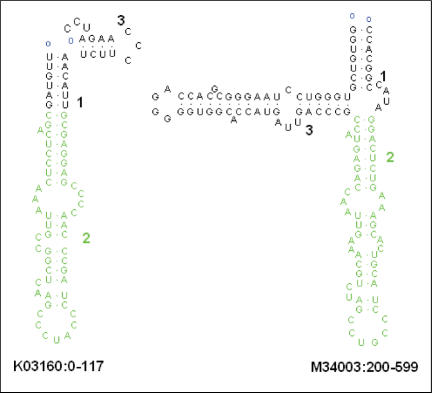
Upon completion of a structure alignment job, RADAR presents the alignment result on a web page where the alignment result can be downloaded to a file on a local machine. In Figure 2, the common region of two RNA secondary structures given in an alignment result is portrayed using RnaViz (19), where the local matches in the two structures are highlighted with the green color. For pairwise structure alignment and constrained structure alignment, RADAR ranks the subject structures in the given set based upon their similarities to the query structure. The top ranked subject structure is most similar to the query structure, with the maximum alignment score. The score diminishes as the quality of the alignment decreases.
Figure 2.
An example showing the common region of two RNA secondary structures where the local matches in the two structures are highlighted with the green color.
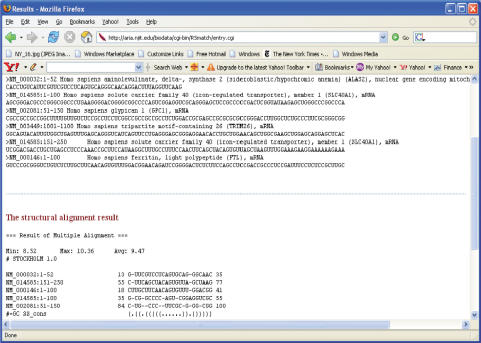
In the case of a multiple structure alignment, RADAR displays the set of RNA structures followed by the best possible multiple structure alignment obtained from them. Inference can be made from looking at this alignment about the presence of a common motif. Such knowledge greatly helps in studying and discovering the functionalities of RNA molecules. Figure 3 shows an example of the output from the RADAR function to predict the consensus structure for a set of RNA sequences. The input sequences are shown on the top of the figure and the consensus structure is shown at the bottom of the figure. In the figure, the consensus structure is an iron regulatory element (IRE) (20) and all the input sequences are known IRE-containing sequences. The IRE motif is displayed as a multiple structure alignment where the alignment shows the positions at which the motif occurs in each input sequence. These positions indicate the offsets within a sequence. For example, in NM_014585:151-250, the motif begins at the 55th position and ends at the 77th position of the sequence. Details regarding the testing and experiments concerning consensus structure prediction are provided in Supplementary Material.
Figure 3.
An example of the output from RADAR's consensus-structure prediction function for a set of RNA sequences. The result shows a group of subsequences from the input that consists of a common structure. Here the common structure is that of the IRE motif (20).
CONCLUSIONS
The RADAR web server provides multiple capabilities for RNA structure alignment data analysis, which includes pairwise structure alignment, multiple structure alignment, constrained structure alignment, database search, clustering and the prediction of a consensus RNA structure from structure alignments for a set of RNA sequences. The web server is implemented in Perl-CGI, rather than SOAP, and hence it requires human–computer interaction.
In future work we plan to apply RADAR to RNA genomes of various organisms to search for motifs in the genomes. The RSmatch algorithm (16) on which RADAR is based is built upon the premise that MFE (minimum free energy) structure prediction for a single sequence is accurate. However, this may not always be true (5, 21), which may impact the accuracy of our method. We plan to carry out an alternative approach similar to the CMfinder method (22) by using blastclust to cluster the RNA sequences at the input, then aligning the resulting clusters using a multiple sequence alignment tool, and finally using KNetFold (7) or a similar tool to predict the structures from the multiple sequence alignments [as outlined in (23)]. This approach would enhance the sensitivity of motif detection in the RNA genomes. To be able to statistically determine the significance of the alignment scores computed by RSmatch, we also plan to provide an e-value for each alignment score.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the anonymous reviewers for their constructive suggestions, which helped to improve the presentation and quality of this article. Funding to pay the Open Access publication charges for this article was provided by NCI-Frederick. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brown V, Ceman S, Peng J, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Wilkinson KD, Keene JD, et al. Identification of mRNAs associated with the fragile X mental retardation protein complex in the brain. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 2.Ford LP, Bagga PS, Wilusz J. The poly (A) tail inhibits the assembly of a 3′ to 5′ exonuclease in an in vitro RNA stability system. Mol. Cell. Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haugen P, Runge HJ, Bhattacharya D. Long-term evolution of the S788 fungal nuclear small subunit rRNA group I introns. RNA. 2004;10:1084–1096. doi: 10.1261/rna.5202704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapiro BA, Bengali D, Kasprzak W, Wu JC. RNA folding pathway functional intermediates: their prediction and analysis. J. Mol. Biol. 2001;312:27–44. doi: 10.1006/jmbi.2001.4931. [DOI] [PubMed] [Google Scholar]
- 5.Gardner PP, Giegerich R. A comprehensive comparison of comparative RNA structure prediction approaches. BMC Bioinformatics. 2004;5:140. doi: 10.1186/1471-2105-5-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shapiro BA, Kasprzak W, Grunewald C, Aman J. Graphical exploratory data analysis of RNA secondary structure dynamics predicted by the massively parallel genetic algorithm. J. Mol. Graph. Modeling. 2006;25:514–531. doi: 10.1016/j.jmgm.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Bindewald E, Shapiro BA. RNA secondary structure prediction from sequence alignments using a network of k-nearest neighbor classifiers. RNA. 2006;12:342–352. doi: 10.1261/rna.2164906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bindewald E, Schneider TD, Shapiro BA. CorreLogo: an online server for 3D sequence logos of RNA and DNA alignments. Nucleic Acids Res. 2006;34:405–411. doi: 10.1093/nar/gkl269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofacker IL. RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proc. Roy. Soc. (London) B. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- 11.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 12.Akmaev VR, Kelley ST, Stormo GD. Phylogenetically enhanced statistical tools for RNA structure prediction. Bioinformatics. 2000;16:501–512. doi: 10.1093/bioinformatics/16.6.501. [DOI] [PubMed] [Google Scholar]
- 13.Gulko B, Haussler D. Proceedings of the 1st Pacific Symposium on Biocomputing. Hawaii, USA: World Scientific in Singapore; 1996. Using multiple alignments and phylogenetic trees to detect RNA secondary structure; pp. 350–367. [PubMed] [Google Scholar]
- 14.Knudsen B, Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas E, Eddy SR. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 1999;285:2053–2068. doi: 10.1006/jmbi.1998.2436. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Wang JTL, Hu J, Tian B. A method for aligning RNA secondary structures and its application to RNA motif detection. BMC Bioinformatics. 2005;6:89. doi: 10.1186/1471-2105-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rijk PD, Wuyts J, Wachter RD. RnaViz2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- 20.Theil EC. The IRE (iron regulatory element) family: structures which regulate mRNA translation or stability. Biofactors. 1993;4:87–93. [PubMed] [Google Scholar]
- 21.Doshi KJ, Cannone JJ, Cobaugh CW, Gutell RR. Evaluation of the suitability of free-energy minimization using nearest-neighbor energy parameters for RNA secondary structure prediction. BMC Bioinformatics. 2004;5:105. doi: 10.1186/1471-2105-5-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao Z, Weinberg Z, Ruzzo WL. CMfinder - a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 23.Freyhult EK, Bollback JP, Gardner PP. Exploring genomic dark matter: a critical assessment of the performance of homology search methods on noncoding RNA. Genome Res. 2007;17:117–125. doi: 10.1101/gr.5890907. [DOI] [PMC free article] [PubMed] [Google Scholar]