Abstract
The DBAli tools use a comprehensive set of structural alignments in the DBAli database to leverage the structural information deposited in the Protein Data Bank (PDB). These tools include (i) the DBAlit program that allows users to input the 3D coordinates of a protein structure for comparison by MAMMOTH against all chains in the PDB; (ii) the AnnoLite and AnnoLyze programs that annotate a target structure based on its stored relationships to other structures; (iii) the ModClus program that clusters structures by sequence and structure similarities; (iv) the ModDom program that identifies domains as recurrent structural fragments and (v) an implementation of the COMPARER method in the SALIGN command in MODELLER that creates a multiple structure alignment for a set of related protein structures. Thus, the DBAli tools, which are freely accessible via the World Wide Web at http://salilab.org/DBAli/, allow users to mine the protein structure space by establishing relationships between protein structures and their functions.
INTRODUCTION
The number of known protein structures deposited in the Protein Data Bank (PDB) has grown exponentially over the years (1). This trend is expected to continue, partly due to the structural genomics efforts (2,3). Currently, there are ∼41 000 protein structures deposited in the PDB, containing ∼88 000 protein chains. These protein structures constitute a structural space that can be mined to facilitate the understanding, assignment and modification of protein function. Previously developed databases for the classification of protein structure domains, such as SCOP [http://scop.mrc-lmb.cam.ac.uk/scop/(4)] or CATH [http://www.cathdb.info (5)], and servers for functional annotation of protein structures, such as ProFunc [http://www.ebi.ac.uk/thornton-srv/databases/ProFunc/ (6,7)], ProKnow [http://www.doe-mbi.ucla.edu/Services/ProKnow (8)] and Phunctioner [http://www.sbg.bio.ic.ac.uk (9)], provide an effective way of describing and annotating the protein structure space. However, none of these servers combine a comprehensive database of protein structural alignments with tools for automatically annotating protein structures.
Here, we describe five tools that aid in the analysis of the data stored in DBAli, our comprehensive relational database of pairwise and multiple structural alignments (10). These tools include (i) the DBAlit program that allows users to input their structure for comparison by MAMMOTH (11) against all chains in the PDB; (ii) the AnnoLite and AnnoLyze programs that annotate a target structure based on its stored relationships to other structures; (iii) the ModClus program that clusters structures by sequence and structure similarities; (iv) the ModDom program that identifies recurrent fragments, including domains, from structure; and (v) an implementation of the COMPARER method (12) in the SALIGN command in MODELLER that creates a multiple structure alignment for a set of related protein structures. The DBAli tools allow users to establish relationships between protein structures and their fragments in a flexible and dynamic manner.
The DBAli database is briefly introduced first. Next, we describe each of the five tools that make use of the structural alignments deposited in DBAli. Finally, we discuss the use of the DBAli tools to analyze a structure determined by the New York Structural Genomics Research Consortium (NYSGXRC).
THE DBALI SERVER
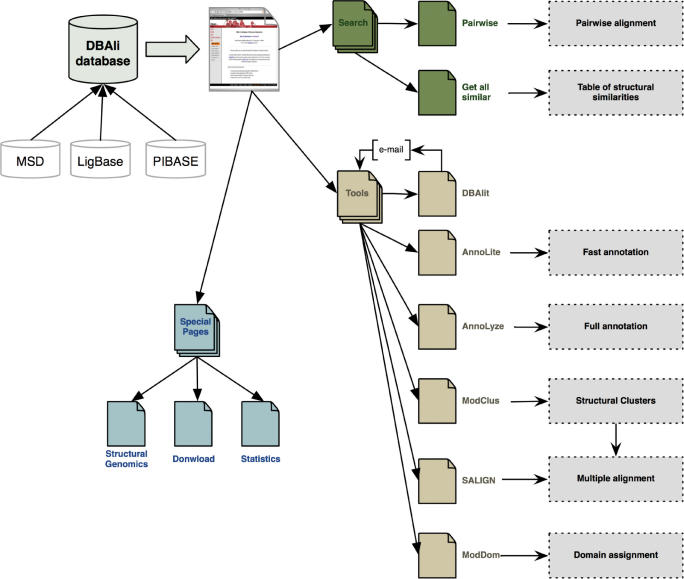
The DBAli server (http://salilab.org/DBAli/) is divided into four main sections: the DBAli database, search pages, tools pages and special pages, each one of which is dedicated to a specific tasks (Figure 1).
Figure 1.
DBAli server organization.
The DBAli database
The DBAli database contains pairwise and multiple structure alignments of protein structures in the PDB. Pairwise alignments are updated weekly and multiple alignments are updated monthly. Currently (January 2007), DBAli contains a total of 86,277 PDB chains in ∼1.38 billion pairwise alignments with a MAMMOTH P-value higher than 2.0 (Table 1). DBAli also stores multiple structure alignments for 11,615 families with 30,900 non-redundant PDB chains representing 86,277 chains in PDB (Table 1). Structural and functional annotations are obtained from our databases LigBase (13) and PIBASE (14) and external databases such as CATH (5), SCOP (4), InterPro (15), PFam (16), EC (17) and GO (18). Cross-links between the external databases is adopted from the MSD database (19) (Table 1).
Table 1.
Internal and external data sources used by the DBAli tools
| Database | Last update | Type of information | Number of entries | Reference |
|---|---|---|---|---|
| DBAli pairwise | January 2007 | Structural alignments | 1,379,352,642 | (10) |
| DBAli multiple | January 2007 | Structural alignments | 11,615 | (10) |
| MSD | November 2006 | PDB chains | 79,170 | (19) |
| CATH | July 2006 | Superfamilies | 2,359 | (5) |
| SCOP | April 2006 | Domains | 94,779 | (4) |
| InterPro | July 2006 | Domains and motifs | 13,057 | (15) |
| PFam | July 2006 | Protein families | 8,376 | (16) |
| EC | November 2006 | Enzymes | 32,137 | (17) |
| GO | July 2006 | Functional terms | 21,017 | (18) |
| LigBase | February 2004 | Protein ligands | 101,359 | (13) |
| PIBASE | September 2004 | Protein interactions | 158,915 | (14) |
The DBAli tools
DBAli incorporates five tools that use the structural relationships in DBAli.
DBAlit
The DBAlit program takes a user input structure in the PDB format and compares it against all chains in the PDB using the MAMMOTH algorithm. On average, a user-input structure is compared against all known protein structure chains in ∼200 min using 10 Xeon CPUs. To preserve the privacy of the coordinates, DBAli generates a random and unique chain identifier that is returned to the user by e-mail. The new chain identifier can then be used to retrieve data from the DBAli database as well as to use the other tools outlined below.
AnnoLite and AnnoLyze
The AnnoLite and AnnoLyze programs (20) annotate the structures processed by DBAli. A given structure is characterized by annotations of related structures in the MODBASE database of comparative models (21), the PIBASE database of structurally defined protein interfaces (14), the LIGBASE database of small molecule binding sites (13) and the MSD database of macromolecular structures (19). The inputs for the annotation programs are a PDB chain code and thresholds for filtering sequence and structure similarities. The AnnoLite program searches the DBAli database for structurally similar proteins and collects their known annotations. Next, a P-value score is calculated for each transferred annotation using a Fisher's exact test for 2 × 2 contingency tables comparing two groups of annotated chains (i.e. the group of similar chains to the query and the group of all annotated chains in the PDB)(22). Currently, AnnoLite annotates the input protein structure with CATH (5) and SCOP (4) fold assignments, EC numbers (17), InterPro entries (15), PFAM families (16) and Gene Ontology codes (23). The accuracy and coverage of AnnoLite were benchmarked with a set of fully annotated 1,879 non-redundant PDB chains. AnnoLite can reliably annotate a structure for all of the functional properties, with the exception of the GO cellular component term. For example, the CATH fold can be recovered for 96% of the dataset with 89% reliability and direct functional annotation with EC numbers and Gene Ontology molecular function codes can be recovered with reliabilities of 81 and 74% for 83 and 88% of the dataset, respectively. Additionally, AnnoLyze inherits ligands from LIGBASE and interacting partners from PIBASE. The output from the two programs provides an automatic annotation of the protein structures.
ModClus
The ModClus program clusters protein structures based on their sequence and structure similarities. The input to ModClus is a list of PDB chain codes. The output is a list of clusters of the input chains. The clustering depends on user-defined thresholds for structure and sequence similarities. ModClus implements a greedy algorithm as follows: (i) the first chain in the list seeds the first cluster; (ii) the next chain is compared by sequence and/or structure to all chains in each of the existing clusters; it either joins the first sufficiently similar cluster or seeds a new cluster if it is not sufficiently similar to all of the chains in any of the other clusters; (iii) the clustering continues with step (ii) until all chains are processed.
ModDom
The ModDom program assigns domain boundaries in a given structure using the superpositions stored in DBAli. The input is a PDB chain code, which is used as a query to identify structurally similar chains in DBAli. ModDom relies on the relationship between recurrent structures and structural units to predict domain boundaries. The program first builds a residue co-occurrence matrix based on structural alignments selected from the DBAli database and then clusters residue co-occurrences to find common fragments in the query protein structure. The ModDom program has been benchmarked for domain assignment with a non-redundant set of protein structures. ModDom assigns 80% of residues identically to the domain assignments in the SCOP database. The user is provided with all domain assignments and their scores, a structural conservation profile based on the retrieved alignments with similar structures, and a JMol window for inspection of the domain assignments.
SALIGN
The SALIGN command, an implementation of the COMPARER program (12) in MODELLER, generates a multiple structure alignment given a list of PDB chain codes. The alignment is displayed through the JMol applet. In addition, an HTML frame presents an easy-to-read sequence alignment corresponding to the structural superposition. The user is provided with options to download the alignment in the HTML or PIR formats and the superposed coordinates in the PDB format.
UTILITY AND DISCUSSION
Target selection strategies for structural genomics have led to the experimental determination of many protein structures whose functions are not yet known. The tools in DBAli can be employed to annotate the functions of such structures, as illustrated by the following example.
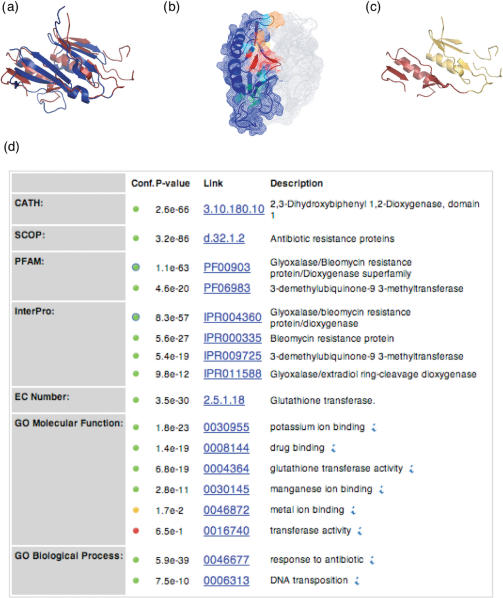
The New York Structural Genomics Research Consortium (NYSGXRC) selected a hypothetical protein from Pseudomonas aeruginosa as a target for structure determination (target T1794). The structure was successfully determined and deposited in the PDB database (code 1u6l, release date 14 December 2004). Searches by PSI-BLAST (24) and threading by GenThreader (25) indicated similarity to the glyoxalase/bleomycin domain (PfamA family PF00903). This domain is found in several proteins including the bleomycin resistance protein and dioxygenases. The DBAli tools confirm and add to these findings. A search for structures similar to chain A of 1u6l (1u6lA) results in 306 related structures. The first annotated hit in the list of similar structures corresponds to a bleomycin resistance protein (PDB code 1xrkB). As a result, the AnnoLyze program (all parameters set to their default values except for minimal sequence identity set to 15%) predicts a binding site on 1u6l that may bind a bleomycin-like ligand. The AnnoLite program predicts that the 1u6lA chain adopts the glyoxalase/bleomycin resistance fold (SCOP code d.32.1.1). ModDom detects that the protein in fact contains two glyoxalase/bleomycin resistance fold domains. In summary, ModDom, AnnoLyze and AnnoLite, adding to the results from sequence-based searches, annotate 1u6lA as a two-domain antibiotic resistance protein and localize a putative binding site for the antibiotic bleomycin (Figure 2).
Figure 2.
Functional annotation of the target T1794 from the NYSGXRC consortium (PDB code 1u6l chain A). (a) The structure closest to the 1v6lA chain with known annotation is an antibiotic inhibitor from Streptoallateichus hindustanus (PDB code 1xrk chain B). It is shown in the ribbon representation in red, superposed on 1u6lA in blue (RMSD of 3.5 Å over 106 residues, sequence identity of 14.3% and MAMMOTH P-value = 13.8). (b) Predicted binding site for bleomycin A2 ligand (residues colored in red and orange: Lys80, Gly81, Cys82, Ser83, Ser85, Ans87, Gln108, Phe115, Trp116, Ser119, Gly121, Thr124, Gly128, Val129, Ala130 and Val133). (c) Domain boundaries of 1u6lA as assigned by ModDom. The protein structure is predicted to have two domains: residues 1–76 (yellow) and residues 77–123 (red). (d) Screen capture of the AnnoLite results for the 1u6lA chain.
CONCLUSIONS
The DBAli database stores a comprehensive and up-to-date comparison of protein structures in the PDB. The data are stored in a MySQL relational database and can be accessed and downloaded via a web server. Several tools have been developed that interact with the data deposited in DBAli, including the DBAlit, AnnoLite and AnnoLyze, ModClus, ModDom, and SALIGN programs. The design of DBAli allows easy cross-referencing to other databases. DBAli already includes links to the MODBASE, LigBase, PIBASE, PDB, CATH, SCOP, PFamA, InterPro, Enzyme and GO databases. Through further integration of new tools and other databases, DBAli and its tools are becoming a valuable resource to the structural biology community. As of January 2007, the DBAli tools have been used by more than 1,700 unique users from 79 different countries worldwide who performed on average ∼600 tasks per month.
AVAILABILITY AND REQUIREMENTS
DBAli is freely available on the Internet at http://salilab.org/DBAli and requires a web browser that is capable of running the JMol applet. The web interface is programmed in PHP and MySQL supports the underlying database.
ACKNOWLEDGEMENTS
We are grateful to Dr Angel Ortiz for the MAMMOTH program. We would also like to thank the JMol team, Erik Bosrup and Stijn van Dongen for JMol, overLib and MCL, respectively. We appreciate valuable comments from Prof Helen M. Berman and Prof. Wayne F. Anderson. We acknowledge funding by The Sandler Family Supporting Foundation, NIH grants GM 62529, GM074929, GM71790 and GM54762, as well as hardware gifts from IBM, Intel, HP and Network Appliance. Funding to pay the Open Access publication charges for this article was provided by Generalitat Valenciana Starting Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burley SK, Almo SC, Bonanno JB, Capel M, Chance MR, Gaasterland T, Lin D, Sali A, Studier FW, et al. Structural genomics: beyond the human genome project. Nat.Genet. 1999;23:151–157. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- 3.Vitkup D, Melamud E, Moult J, Sander C. Completeness in structural genomics. Nat. Struct. Biol. 2001;8:559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- 4.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32(Database issue):D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laskowski RA, Watson JD, Thornton JM. From protein structure to biochemical function? J. Struct. Funct. Genomics. 2003;4:167–177. doi: 10.1023/a:1026127927612. [DOI] [PubMed] [Google Scholar]
- 8.Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13:121–130. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Pazos F, Sternberg MJ. Automated prediction of protein function and detection of functional sites from structure. Proc. Natl Acad. Sci. USA. 2004;101:14754–14759. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marti-Renom MA, Ilyin VA, Sali A. DBAli: a database of protein structure alignments. Bioinformatics. 2001;17:746–747. doi: 10.1093/bioinformatics/17.8.746. [DOI] [PubMed] [Google Scholar]
- 11.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sali A, Blundell TL. Definition of general topological equivalence in protein structures. A procedure involving comparison of properties and relationships through simulated annealing and dynamic programming. J. Mol. Biol. 1990;212:403–428. doi: 10.1016/0022-2836(90)90134-8. [DOI] [PubMed] [Google Scholar]
- 13.Stuart AC, Ilyin VA, Sali A. LigBase: a database of families of aligned ligand binding sites in known protein sequences and structures. Bioinformatics. 2002;18:200–201. doi: 10.1093/bioinformatics/18.1.200. [DOI] [PubMed] [Google Scholar]
- 14.Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 15.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bradley P, Bork P, Bucher P, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33(Database Issue):D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velankar S, McNeil P, Mittard-Runte V, Suarez A, Barrell D, Apweiler R, Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33(Database Issue):D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marti-Renom MA, Rossi A, Al-Shahrour F, Davis FP, Pieper U, Dopazo J, Sali A. The AnnoLite and AnnoLyze programs for comparative annotation of protein structures. BMC Bioinformatics. 2007 doi: 10.1186/1471-2105-8-S4-S4. (Suppl 4):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 23.Camon E, Magrane M, Barrell D, Binns D, Fleischmann W, Kersey P, Mulder N, Oinn T, Maslen J, et al. The Gene Ontology Annotation (GOA) project: implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 2003;13:662–672. doi: 10.1101/gr.461403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DT. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]