Abstract
OPTIMIZER is an on-line application that optimizes the codon usage of a gene to increase its expression level. Three methods of optimization are available: the ‘one amino acid–one codon’ method, a guided random method based on a Monte Carlo algorithm, and a new method designed to maximize the optimization with the fewest changes in the query sequence. One of the main features of OPTIMIZER is that it makes it possible to optimize a DNA sequence using pre-computed codon usage tables from a predicted group of highly expressed genes from more than 150 prokaryotic species under strong translational selection. These groups of highly expressed genes have been predicted using a new iterative algorithm. In addition, users can use, as a reference set, a pre-computed table containing the mean codon usage of ribosomal protein genes and, as a novelty, the tRNA gene-copy numbers. OPTIMIZER is accessible free of charge at http://genomes.urv.es/OPTIMIZER.
INTRODUCTION
Gene expression levels depend on many factors, such as promoter sequences and regulatory elements. One of the most important factors is the adaptation of the codon usage of the transcript gene to the typical codon usage of the host (1). Therefore, highly expressed genes in prokaryotic genomes under translational selection have a pronounced codon usage bias. This is because they use a small subset of codons that are recognized by the most abundant tRNA species (2). The force that modulates this codon adaptation is called translational selection and its strength is important in fast-growing bacteria (3,4). If a gene contains codons that are rarely used by the host, its expression level will not be maximal. This may be one of the limitations of heterologous protein expression (5) and the development of DNA vaccines (6). A high number of synthetic genes have been re-designed to increase their expression level. The Synthetic Gene Database (SGDB) (7) contains information from more than 200 published experiments on synthetic genes. In the design process of a nucleic acid sequence that will be inserted into a new host to express a certain protein in large amounts, codon usage optimization is usually one of the first steps (5). Codon usage optimization basically involves altering the rare codons in the target gene so that they more closely reflect the codon usage of the host without modifying the amino acid sequence of the encoded protein (5). The information usually used for the optimization process is therefore the DNA or protein sequence to be optimized and a codon usage table (which we call the reference set) of the host.
Here we present a new web server, called OPTIMIZER, for codon usage optimization focused on the heterologous, or even homologous, gene expression in bacterial hosts. OPTIMIZER allows three optimization methods and uses several valuable, new reference sets. OPTIMIZER can therefore be used to optimize the expression level of a gene, to assess the adaptation of alien genes inserted into a genome (8), or to design new genes from protein sequences. The server is freely available at http://genomes.urv.es/OPTIMIZER. It has been running since July 2005 and it is updated twice a year with new features and reference sets.
PROGRAM OVERVIEW
Implementation and input data
OPTIMIZER is an on-line application and its methods are implemented in PHP (hypertext pre-processor) programming language. The pre-calculated reference tables are stored into a MySQL database. The data input and the selection of the server options have been organized in four steps. These steps are: (1) Input the sequence to be optimized. DNA or protein sequences can be used, although further steps are slightly different depending on whether a DNA or protein sequence has been input. (2) Input the reference set. Users can insert a codon usage table in a variety of formats, including tables from the Codon Usage Database (9), or they can choose between 153 pre-computed codon usage tables for ribosomal protein genes or a group of highly expressed genes from prokaryotic genomes under translational selection. Users can also choose a reference set consisting of the tRNA gene-copy numbers. (3) Choose the genetic code. (4) Choose the method to be used in the optimization process. Depending on the type of sequence introduced (DNA or protein) and the reference set chosen, different optimization methods are available (see below for a description of the optimization methods).
Calculation of the reference sets
One of the main features of the OPTIMIZER server is that it contains a series of pre-computed reference sets that can be used in the optimization process. These reference sets can be a table containing the codon usage of the host (or the codon usage of a group of genes, such as the group of highly expressed genes) or, as a novelty, the number of tRNA gene copies predicted with the tRNA-scan software (10). The pre-computed reference sets available in the server are from more than 150 prokaryotic genomes that are under a strong translational selection. The codon usage reference tables available for these genomes contain the mean codon usage of genes that encode ribosomal proteins or a group of highly expressed genes. Although the optimization process can be carried out using the mean codon usage of the host organism as a reference set, if the aim of the optimization process is to increase the expression level of a gene, it is preferable to use the codon usage of a group of highly expressed genes. The mean codon usage of bacteria is highly influenced by mutational bias (i.e. their G + C content). The optimal codons (those most frequently used in highly expressed genes) are usually those that agree with the mutational bias (i.e. G- or C-ending codons for G + C-rich organisms). However, the optimal codons are not always in agreement with mutational bias. For example, in the amino acids that are coded by only two synonymous codons ending in C or T, the C-ending codon is usually preferred, independently of the mutational bias (3). Therefore, using the mean codon usage of a genome may cause the wrong choice of optimal codons.
A new feature of the OPTIMIZER server is that it can use tRNA gene-copy numbers as a reference set for the optimization process. If the codon usage bias of highly expressed genes is caused by differences in tRNA gene-copy numbers, why not use this information for the optimization process? At present, information about tRNA gene-copy numbers is used in the OPTIMIZER server only with the ‘one amino acid–one codon’ optimization method (for a complete description of the methods available, see the ‘Optimization methods’ section below).
Evaluation of which bacterial genomes are under translational selection
Not all prokaryotic species are under translational selection (4,11). It would be pointless to optimize the codon usage of a gene in order to increase its expression level in a species such as Helicobacter pylori, which is not under translational selection (i.e. in which the highly expressed genes do not have a different pattern of codon usage from the other genes of their genome) (12). Traditionally, correspondence analysis of the relative synonymous codon usage of all genes from a genome has been used to detect whether a genome is under translational selection (13). In genomes under translational selection, the ribosomal protein genes and other highly expressed genes form a cluster in the correspondence analysis plot, which confirms that highly expressed genes have a different codon usage from the other genes of a genome. This is the method we have used to detect which bacterial species are under translational selection. For each bacterial complete genome available, we made a correspondence analysis using the Relative Synonymous Codon Usage (RSCU) values of all the genes of a genome. To automate the analysis of the correspondence plots, we analyzed the position of the ribosomal protein genes (expected to be highly expressed genes) along the four principal axes obtained in the correspondence analysis. If a genome is under translational selection, ribosomal proteins and other highly expressed genes will show a codon usage bias and they will form a cluster in the correspondence plot. To make the prediction of translational selection, we checked whether the mean position of the ribosomal protein genes along any of the four principal axes was significantly different (evaluated with a t-test) from the mean position of the other genes of their genome. To check our predictions, we also visually inspected the correspondence plots (correspondence analysis plots are available from the homepage of the server) and analyzed the metabolic function of the predicted highly expressed genes obtained. Analysis of 334 prokaryotic genomes revealed that 153 genomes (the total number of different species and genera was 108 and 63, respectively) were under a strong translational selection. These genomes were then used to calculate the pre-computed reference sets.
Prediction of highly expressed genes
The predicted highly expressed genes were obtained using an iterative algorithm that we have developed. This algorithm uses the group of genes that encode ribosomal proteins as a seed and, through a series of iterations, define a group of putative highly expressed genes. This algorithm works as follows:
Using the functional annotation, gene names or COG families, genes that encode ribosomal proteins are detected. Using the codon usage of these genes as a reference set, the Codon Adaptation Index (CAI), (14), at this stage namely CAIrp (15), is calculated for each gene of a genome.
Using now the group of genes with the highest CAI values as a reference set, the CAI for all genes is recalculated. This process is repeated until a homogeneous group is reached, i.e. when the group of genes with the highest CAI values in one iteration is the same as the group in the next iteration.
To provide further support for our predictions, we analyzed the metabolic functions of the putative highly expressed genes. As expected, ribosomal proteins and other expected highly expressed genes (16) were found in the final group of predicted highly expressed genes. To check our algorithm, we also analyzed species not under translational selection. With these species, either the algorithm never ended or the final group of genes had a high codon usage bias but was not related to their expression level. In this situation, neither ribosomal protein genes nor genes expected to have a high expression were included in the final group of genes with a codon usage bias. Our method is similar to the one developed by Carbone and co-workers (17). However, these authors used all the genes of an organism as the initial reference set, whereas we used ribosomal protein genes.
Optimization methods
The OPTIMIZER server provides three methods for optimizing the codon usage of the query sequence. In the first method, the ‘one amino acid–one codon’ method, all the codons that encode the same amino acid are substituted by the most commonly used synonymous codon in the reference set. However, this approach has several drawbacks: for example, translational errors may be made due to an imbalanced tRNA pool and it is impossible to avoid repetitive elements or cleavage sites of restriction enzymes (5,18). To overcome these drawbacks, a second method, which we call the ‘guided random’ method, can be used. This method consists of a Monte Carlo algorithm that selects codons at random based on the frequencies of use of each codon in the reference set. The third method, which we call the ‘customized one amino acid–one codon’ method, is an intermediate method in which users choose how many of the 59 codons (if the standard genetic code has been selected) will be optimized with the ‘one amino acid–one codon’ approach. ‘Rare codons’ (i.e. the least used codons in the reference set) are the first codons changed with this approach. The aim of this third method is to maximize the optimization by making the fewest changes in the query sequence.
If the input sequence is a protein, it can be back-translated to DNA using the ‘one amino acid–one codon’ or the ‘guided random’ approach. If the ‘one amino acid–one codon’ approach is selected, the protein sequence can be back-translated to DNA using codons with the highest G + C or A + T content, or codons defined by Archetti (19) that minimize mutation errors.
Outputs
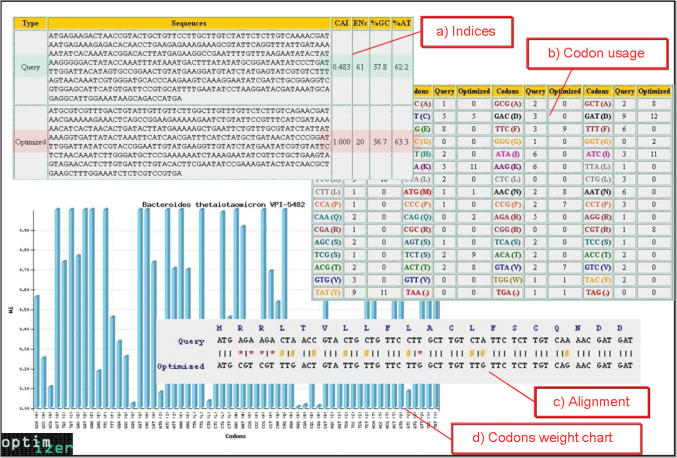
Two indices, CAI and ENc (effective number of codons), are used to measure the optimization process. CAI measures the similarity between the codon usage of a gene and the codon usage of a reference group of genes (14). Its values range from 0 (when the codon usage of a sequence and that of the reference set are very different) to 1 (when both codon usages are the same). This index is the most effective of all codon bias measures for predicting gene expression levels (12,20). The second index is ENc, which is a measure of codon usage bias (21). Its values range from 20 (if only one codon per amino acid is used) to 61 (if all synonymous codons are used equally). Because highly expressed genes usually use the minimal subset of codons that are recognized by the most abundant tRNA species, their ENc values are expected to be low. Figure 1 shows some of the outputs provided by the optimization of a DNA sequence: for example, the query and optimized sequences and an alignment between them, a chart of the relative frequencies of each codon of the reference set and a codon usage table of the query and optimized sequences. In addition, the OPTIMIZER server has options for viewing or avoiding the cleavage sites of the selected restriction enzymes (22) and for splitting the optimized sequence into several overlapping oligonucleotides for the construction of a synthetic gene.
Figure 1.
Outputs provided from the optimization of a DNA sequence: (a) the optimized and query sequences and the indices (CAI, ENc and%G + C) for evaluating the optimization process, (b) codon usage tables of the query and optimized sequences, (c) query and optimized sequence alignment to show changes in nucleotides (transitions or transversions) and (d) graphical view of the codon weight chart.
Comparison with other servers and programs
Table 1 shows a comparison of several public web servers and stand-alone applications that allow some kind of codon optimization. ‘GeneDesign’ (23), ‘Synthetic Gene Designer’ (24) and ‘Gene Designer’ (18) are packages that provide a platform for synthetic gene design, including a codon optimization step. Other programs, such as DNAWorks (25) and GeMS (26), focus more on the process of oligonucleotide design for synthetic gene construction. The stand-alone application INCA provides an array of features, including now codon optimization, which are useful for analyzing synonymous codon usage in whole genomes (27). JCAT (28), ‘Codon optimizer’ (29), UpGene (30) and the server presented here focus on the codon optimization process. Although each server and application has its own features, all of them have several features in common. Most offer several options for the input of the codon usage reference set. One of these options is the possibility of using the tables from the Codon Usage database (9). Usually, a limited number of pre-computed tables of codon usage are available to be used as a reference set in the optimization process. In addition, not all of the available pre-computed reference sets correspond to a group of highly expressed genes (the proper reference set needed to optimize for increasing gene expression level). Though most of the programs and servers use a group of highly expressed genes from E. coli as a pre-computed reference set, only the ‘Synthetic Gene Designer’ and ‘GeneDesign’ servers provide a pre-computed group of highly expressed genes for 11 and 4 organisms, respectively. The exception is the JCAT web server, which offers pre-computed tables of predicted highly expressed genes from more than 200 bacterial species. However, this server uses the method of Carbone et al. (17) to predict a group of genes with a biased codon usage. These groups of genes do not always correspond to a group of highly expressed genes because not all bacterial species are under translational selection (11,17). The high number of pre-computed codon usage tables from bacteria and archaea that are not under translational selection available in JCAT therefore creates some confusion. The OPTIMIZER server presented here provides the most pre-computed codon usage tables for use as a reference set. The OPTIMIZER server provides pre-computed tables for more than 150 prokaryotic genomes that are under strong translational selection. In addition, two groups of genes are available in each reference set: a group of highly expressed genes predicted using a new prediction algorithm and the group of ribosomal protein genes. OPTIMIZER is the only server or stand-alone application that introduces a new kind of reference set such as information about the number of copies of tRNA genes for all the species included in the server. With regard to the methods for codon usage optimization available in each server or program, the first programs developed used only the ‘one amino acid–one codon’ approach. More recent programs and servers now include further methods to create some codon usage variability. This variability reflects the codon usage variability of natural highly expressed genes and enables additional criteria to be introduced (such as the avoidance of restriction sites) in the optimization process. The OPTIMIZER server presented here provides three methods of codon optimization: a complete optimization of all codons, an optimization based on the relative codon usage frequencies of the reference set that uses a Monte Carlo approach (similar to methods from other programs and servers) and a novel approach designed to maximize the optimization with the minimum changes between the query and optimized sequences. Finally, note that only the ‘Synthetic Gene Designer,’ INCA and OPTIMIZER allow users to choose a non-standard genetic code.
Table 1.
Comparison of OPTIMIZER with other similar freely available web servers and softwares
| Name | Methods | Genetic code | Reference set | Reference |
|---|---|---|---|---|
| Web servers | ||||
| OPTIMIZER | – One amino acid–one codon | Multiple | – HEG from >150 bacterial genomes under TS | This article |
| – Guided Random (Monte Carlo algorithm)b | – RPG | |||
| – Customized one amino acid–one codon | – tGCN | |||
| – Codon usage database | ||||
| – Defined by users | ||||
| JCAT | – One amino acid–one codon | Standard | – HEG from >200 bacterial genomes | 28 |
| – Defined by users | ||||
| Synthetic Gene Designer (SGD) | – One amino acid–one codona | Multiple | – HEG from six bacterial genomes | 24 |
| – Selective optimizationa | – Codon usage database | |||
| – Probabilistic optimizationa,b | – Defined by users | |||
| DNAWorks | – Use of the two highest frequency codons | Standard | – HEG from E. coli | 25 |
| – Random | – Codon usage tables for 10 species | |||
| – Codon usage database | ||||
| – Defined by users | ||||
| GeneDesign | – One amino acid–one codon | Standard | – HEG from four species | 23 |
| – The next most optimal algorithm | – Defined by users | |||
| – The most different algorithm | ||||
| – Random | ||||
| Stand-alone applications | ||||
| Gene Designer | – One amino acid–one codon | Standard | – HEG from E. coli | 18 |
| – Monte Carlo algorithmb | – Codon usage tables for 25 species | |||
| – Codon usage database | ||||
| – Defined by users | ||||
| Codon optimizer | – One amino acid–one codon | Standard | – HEG for several bacterial species | 29 |
| – Defined by users | ||||
| INCA 2.1 | – One amino acid–one codon | Multiple | – Mean codon usage of a whole genome or selection of any group of genes | 27 |
| UPGene | – One amino acid–one codon | Standard | – Eukaryotic, bacteria, yeast, plant and worm predefined codon usage frequency tables | 30 |
| – Defined by users | ||||
| GeMS | – Monte Carlo algorithmb | Standard | – Codon usage database | 26 |
| – Defined by users |
Abbreviations used: HEG, codon usage of predicted highly expressed genes; RPG, codon usage of ribosomal protein genes; tGCN, tRNA gene-copy number; TS, translational selection.
aIt uses an ‘optimality factor,’ defined as a scaling factor, to control the optimality of codon usage. Higher values of this factor mean low CAI values and less optimized and more random codon usage.
bThese methods are essentially the same. They use the relative codon usage frequencies of the reference set as the relative probability that each codon will be used in the optimization process.
CONCLUSIONS
OPTIMIZER is a new codon optimization web server focused on maximizing the gene expression level through the optimization of codon usage. It has unique features, such as a novel definition of a group of highly expressed genes from more than 150 prokaryotic species under translational selection, and the possibility of using information on tRNA gene-copy numbers in the optimization process. OPTIMIZER provides several pre-computed tables to specify a reference set and combines three different methods of codon optimization. The OPTIMIZER server can be used to optimize the expression level of a gene in heterologous gene expression or to design new genes that confer new metabolic capabilities in a given species.
ACKNOWLEDGEMENTS
This work has been financed by project BIO2003-07672 of the Spanish Ministry of Science and Technology. We thank Kevin Costello and John Bates of the Language Service of the Rovira i Virgili University for their help in writing the manuscript and two anonymous referees for their helpful comments. Funding to pay the Open Access publication charges for this article was provided by project BIO2003-07672 of the Spanish Ministry of Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lithwick G, Margalit H. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res. 2003;13:2665–2673. doi: 10.1101/gr.1485203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 3.Rocha EP. Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Ivory C, Chadee K. DNA vaccines: designing strategies against parasitic infections. Genet. Vaccines Ther. 2004;2:17. doi: 10.1186/1479-0556-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu G, Zheng Y, Qureshi I, Zin HT, Beck T, Bulka B, Freeland SJ. SGDB: a database of synthetic genes re-designed for optimizing protein over-expression. Nucleic Acids Res. 2007;35:D76–D79. doi: 10.1093/nar/gkl648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Vallve S, Guzman E, Montero MA, Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willenbrock H, Friis C, Friis AS, Ussery DW. An environmental signature for 323 microbial genomes based on codon adaptation indices. Genome Biol. 2006;7:R114. doi: 10.1186/gb-2006-7-12-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry I, Sharp PM. Predicting gene expression level from codon usage bias. Mol. Biol. Evol. 2007;24:10–12. doi: 10.1093/molbev/msl148. [DOI] [PubMed] [Google Scholar]
- 13.Perriere G, Thioulouse J. Use and misuse of correspondence analysis in codon usage studies. Nucleic Acids Res. 2002;30:4548–4555. doi: 10.1093/nar/gkf565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharp PM, Li WH. The codon adaptation index – a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura Y, Tabata S. Codon-anticodon assignment and detection of codon usage trends in seven microbial genomes. Microb. Comp. Genomics. 1997;2:299–312. doi: 10.1089/omi.1.1997.2.299. [DOI] [PubMed] [Google Scholar]
- 16.Karlin S, Mrazek J. Predicted highly expressed genes of diverse prokaryotic genomes. J. Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone A, Zinovyev A, Kepes F. Codon adaptation index as a measure of dominating codon bias. Bioinformatics. 2003;19:2005–2015. doi: 10.1093/bioinformatics/btg272. [DOI] [PubMed] [Google Scholar]
- 18.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Archetti M. Selection on codon usage for error minimization at the protein level. J. Mol. Evol. 2004;59:400–415. doi: 10.1007/s00239-004-2634-7. [DOI] [PubMed] [Google Scholar]
- 20.Goetz RM, Fuglsang A. Correlation of codon bias measures with mRNA levels: analysis of transcriptome data from Escherichia coli. Biochem. Biophys. Res. Commun. 2005;327:4–7. doi: 10.1016/j.bbrc.2004.11.134. [DOI] [PubMed] [Google Scholar]
- 21.Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 22.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE – restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson SM, Wheelan SJ, Yarrington RM, Boeke JD. GeneDesign: rapid, automated design of multikilobase synthetic genes. Genome Res. 2006;16:550–556. doi: 10.1101/gr.4431306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu G, Bashir-Bello N, Freeland SJ. The synthetic gene designer: a flexible web platform to explore sequence manipulation for heterologous expression. Protein Expr. Purif. 2006;47:441–445. doi: 10.1016/j.pep.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayaraj S, Reid R, Santi DV. GeMS: An advanced software package for designing synthetic genes. Nucleic Acids Res. 2005;33:3011–3016. doi: 10.1093/nar/gki614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Supek F, Vlahovicek K. INCA: Synonymous codon usage analysis and clustering by means of self-organizing map. Bioinformatics. 2004;20:2329–2330. doi: 10.1093/bioinformatics/bth238. [DOI] [PubMed] [Google Scholar]
- 28.Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuglsang A. Codon optimizer: a freeware tool for codon optimization. Protein Expr. Purif. 2003;31:247–249. doi: 10.1016/s1046-5928(03)00213-4. [DOI] [PubMed] [Google Scholar]
- 30.Gao W, Rzewski A, Sun H, Robbins PD, Gambotto A. UpGene: Application of a web-based DNA codon optimization algorithm. Biotechnol. Prog. 2004;20:443–448. doi: 10.1021/bp0300467. [DOI] [PubMed] [Google Scholar]