Abstract
The fastSCOP is a web server that rapidly identifies the structural domains and determines the evolutionary superfamilies of a query protein structure. This server uses 3D-BLAST to scan quickly a large structural classification database (SCOP1.71 with <95% identity with each other) and the top 10 hit domains, which have different superfamily classifications, are obtained from the hit lists. MAMMOTH, a detailed structural alignment tool, is adopted to align these top 10 structures to refine domain boundaries and to identify evolutionary superfamilies. Our previous works demonstrated that 3D-BLAST is as fast as BLAST, and has the characteristics of BLAST (e.g. a robust statistical basis, effective search and reliable database search capabilities) in large structural database searches based on a structural alphabet database and a structural alphabet substitution matrix. The classification accuracy of this server is ∼98% for 586 query structures and the average execution time is ∼5. This server was also evaluated on 8700 structures, which have no annotations in the SCOP; the server can automatically assign 7311 (84%) proteins (9420 domains) to the SCOP superfamilies in 9.6 h. These results suggest that the fastSCOP is robust and can be a useful server for recognizing the evolutionary classifications and the protein functions of novel structures. The server is accessible at http://fastSCOP.life.nctu.edu.tw.
INTRODUCTION
As protein structures become increasingly available and structural genomics provide structural models in a genome-wide strategy (1), proteins with unassigned functions are accumulating and the number of protein structures in the Protein Data Bank (PDB) is rapidly rising (2). The evolutionary classification databases, such as SCOP (3) and CATH (4), are valuable resources for understanding protein functions, structural similarity and evolutionary relationships. However, these two widely used databases are updated intermittently using manual and semi-automated methods. This current structure–function gap clearly reveals the need for powerful automated methods to classify protein domains based on their tertiary structures and is important in producing manually tuned classification databases.
Many automatic domain classification approaches have been developed to determine similar structures and structural classification (5,6) of a query structure. Protein sequence database search tools, such as BLAST (7), PSI-BLAST and Superfamily (5), are useful computational tools. However, these tools are commonly unreliable in detecting remote homologous relationships that are indicated by such structural alignment tools as DALI (8), MAMMOTH (9) and SSM (10). Structural alignment tools typically take several seconds to align two known structures. At this speed, about one day is required to compare a single protein structure with those in PDB. SCOPmap (6), which is computationally more expensive, combines sequence and structural information for SCOP superfamily assignment.
Recently, we have proposed a fast and efficient tool, called 3D-BLAST (11), to quickly search similar structures. This tool is as fast as BLAST and provides the statistical significance (E-value) of an alignment to indicate the reliability of a structure. 3D-BLAST outperformed fast structural search methods (TOPSCAN (12) and YAKUSA (13)) and approached the performance of detailed structural alignment approaches (CE (14) and MAMMOTH (9)). 3D-BLAST is rapid and accurate in scanning a large protein structural database, and is useful in an initial scan for similar protein structures, which can be refined using detailed structural comparison methods. However, several factors that deteriorate 3D-BLAST's performance are (i) 3D-BLAST may have made minor shifts in aligning two local segments with similar letters, because the structural alphabet do not consider actual Euclidean distances, (ii) the E-values of the hit proteins are insignificant and (iii) the query is a multiple-domain protein (11).
This work presents an automated server (fastSCOP), which integrates a fast structure database search tool (3D-BLAST) and a detailed structural alignment tool (MAMMOTH), to recognize SCOP domains and SCOP superfamilies of a query structure. MAMMOTH provided the Z-score and root-mean-square deviation (RMSD) of the Cα atom positions of the aligned residues between the query structure and the hit structure according to the Euclidean distance between corresponding residues rather than the distance between amino acid ‘types’ used in sequence alignments. The classification accuracy of this server is 98% for 464 single-domain queries and 122 multiple-domain queries. To combine 3D-BLAST and MAMMOTH is able to reduce the ill effects of 3D-BLAST to improve the assignment accuracy. After a query structure is assigned to a superfamily, this server is able to provide both multiple sequence alignments and multiple structural alignments of the selected members in a SCOP superfamily.
METHOD AND IMPLEMENTATION
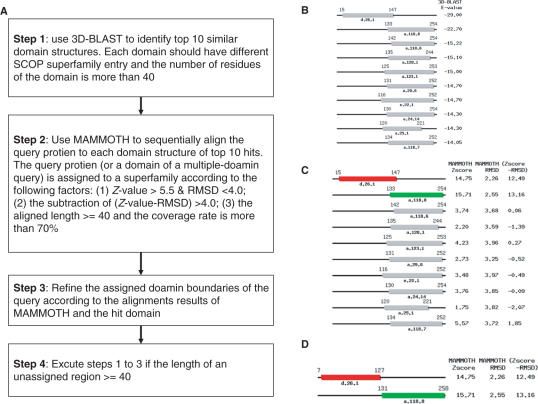
Figure 1 presents an overview of the fastSCOP server for rapidly recognizing SCOP domains and SCOP superfamilies. This sever uses 3D-BLAST to scan quickly the SCOP 1.71 database and selected the top 10 hit domain structures, which are associated with different SCOP superfamily entries (Figure 1B). MAMMOTH was then adopted to align sequentially the query structure with each structure of the top 10 structures to refine the domain boundaries and to recognize SCOP superfamilies (Figure 1C and D). Our previous work (11) demonstrated that 3D-BLAST required ∼1.4 s to scan the structural domains in SCOP 1.69 and was 16 990 and 1413 times faster than CE (14) and MAMMOTH, respectively. These two detailed structural alignment tools perform similarly on the test set; MAMMOTH was ∼12 times faster than CE. The SCOP 1.71 database (October 2006) has 75 930 domains that are derived from 27 599 PDB entries (18 January 2005). The numbers of folds, superfamilies and families are 971, 1589 and 3004, respectively. 3D-BLAST requires structural alphabet sequence databases (SADB) for fast scanning a protein structural database. In this work, we created an SADB derived from known domain structures (12 927 domains) in SCOP1.71 with <95% identity to each other based on the (κ, α) plot (11).
Figure 1.
Overview of the fastSCOP server for SCOP domain recognition and superfamily assignment. The fastSCOP has (A) four main steps, including (B) 3D-BLAST for scanning structural database, (C) MAMMOTH for detailed structural alignment and (D) domain boundary refinement and reassignment.
The fastSCOP server performs four main steps to identify the SCOP domains and superfamilies. First, 3D-BLAST was adopted to identify the similar structures (hit SCOP domains), which are ordered by E-value, of a query structure from an SADB database (Figure 1B). 3D-BLAST is the first tool to provide fast search of a protein structural database using the BLAST, which searches on a SADB database with a structural alphabet substitution matrix (SASM) (11). The fastSCOP then selected the top 10 hit domains that have different SCOP superfamily entries. Based on the structural alphabet alignments between the query and hit SCOP domains, this sever can identify multiple domains if a multiple-domain structure is queried. For each hit domain, the aligned length should be more than 40 residues and the overlap of two neighboring hit domains should be <10% of the query protein.
After the 10 ten hit SCOP domains were identified, this server applied MAMMOTH to align sequentially the query structure with each structure of these hit domains, ordered by E-value. For each structural alignment, MAMMOTH yielded the Z-score and RMSD of the Cα atom positions of the aligned residues between the query structure and the hit structure (Figure 1C). The query structure (or one domain of a multiple-domain protein) was assigned to a SCOP superfamily when the pair-structure alignment satisfied the following criteria: (i) the Z-score exceeds 5.5; (ii) the RMSD value is <4 Å; (iii) the value of (Z-score –RMSD) exceeds 4.0 and (iv) the number of the aligned residues exceeds 40 and the coverage rate between the query protein (domain) and hit domain exceeds 75%. In the third step, the fastSCOP refined the boundaries (the start and end positions) of the assigned domain according to the aligned regions and the sequence length of the hit domain (Figure 1D). Finally, the fastSCOP executed steps 1–3 when the length of the unassigned region of the query structure was more than 40 residues.
Input, output and options
The fastSCOP server can identify the structural domains and determine the evolutionary classification of a query structure from evolutionary classification databases. Users input a PDB code with a protein chain (e.g. 2IF4-A). When the query structure is a new protein structure, the fastSCOP server enables users to input the structure file in PDB format.
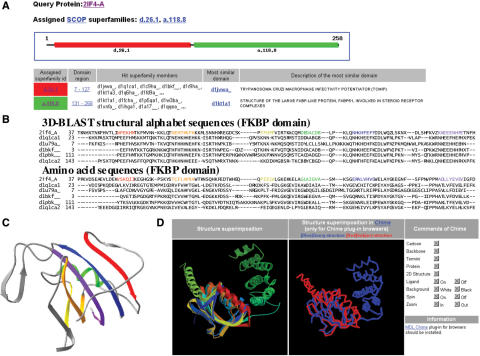
This server typically yielded structural domains and the SCOP superfamilies of a query structure in an average of 6 s (Figure 2A). The server can present the members of the assigned SCOP superfamily and provide both multiple sequence alignments and multiple structural alignments (Figure 2B) based on users’ requirements. The multiple sequence alignments were mapped directly from the aligned results of structural alignments. The aligned structures are visualized in PNG format in MolScript and Raster3D packages (Figure 2C and D). The server allows a user to download the aligned structure coordinates in PDB format.
Figure 2.
Evolutionary superfamily assignment and structural alignment of the fastSCOP server using the structure of multi-domain immunophilin (AtFKBP42) from Arabidopsis thaliana (PDB code 2IF4-A) as the query. (A) The assigned SCOP superfamilies are the FKBP-like domain (SCOP entry d.26.1) and the TPR domain (SCOP entry a.118.8). (B) Multiple structural alphabet and amino acid sequences alignments of FKBP-like domain between the query protein and five homologous proteins. The aligned secondary structures are represented as a continuous color spectrum from red through orange, yellow, green and blue to violet. The color is mapped to (C) the structure of the FKBP-like domain. (D) Structural alignments between the FKBP-like domain of the query protein and that of the homologous protein (PDB code 1Q1C-A).
Example analysis
Figure 2 shows a fastSCOP result with multi-domain immunophilin (AtFKBP42) from Arabidopsis thaliana (PDB code 2IF4-A) (15) as the query structure. The release date of this protein is 31, October 2006, and this protein has not been recorded in SCOP. As shown in Figure 2A, the fastSCOP recognized two domains and their SCOP superfamilies, which are the FKBP-like superfamily (SCOP entry d.26.1) and the TPR-like superfamily (SCOP entry a.118.8) for this query. The FKBP domain (Figure 2C) of AtFKBP42 consists of a six-stranded anti-parallel β-sheet, wrapped around a short α-helix, and is similar to those of FKBP52 (PDB code 1Q1C-A) (16), FKBP 25 (PDB code 1PBK) (17), FKBP 13 (PDB code 1U79-A) (18) and FKBP 12 (PDB code 1BKF) (19). The FKBP domain has been demonstrated to interact with plasma membrane-localized ABC transporters AtPGP1 and AtPGP, which directly mediate cellular auxin efflux (20). The TPR domain of AtFKBP42 is completely helical and binds to AtHSP90, which is critical to plant development and phenotypic plasticity (21,22).
After the structural domains and evolutionary superfamilies were recognized, the fastSCOP server allowed users to browse similar structures of these superfamilies. Using this AtFKBP42 as a query, the server can identify 13 and 17 similar structures of the FKBP-like domain and TPR domain, respectively. Figure 2B illustrates the multiple amino acid sequence alignment and structural alphabet alignment between AtFKBP42 and five FKBP-like homologous proteins, including FKBP52, FKBP 25, FKBP 13 and FKBP 12. The aligned secondary structures are represented as a continuous color spectrum from red through orange, yellow, green and blue to violet (Figure 2B and C). The structural alphabets were strongly conserved in areas of the secondary structures, which are β-strands (represented by structural alphabets E, F, H, K and N) or α-helices (represented by structural alphabets A, Y, B, C and D). These results reveal that the structural alphabet sequences are much better conserved than the amino acid sequences, which result explains why 3D-BLAST detected these distantly related proteins (11).
RESULTS
A query protein set, SCOP-586 (Table 1), was selected to evaluate the utility of the fastSCOP server for recognizing the structural domains and evolutionary superfamilies of a query structure. The SCOP-586 query set has 464 single-domain proteins and 122 multiple-domain proteins that are in SCOP 1.69 but not in SCOP 1.67, and the search database was SCOP 1.67 (11 001 structures). Among the 122 multiple-domain queries, 104 proteins have two domains, 14 have three domains and 4 have more than four domains. The total number of domains is 272 in the multiple-domain query set and the total number of domains in the SCOP-586 is 736.
Table 1.
Accuracy of evolutionary superfamily assignment and average execution time of fastSCOP, 3D-BLAST and MAMMOTH on 586 queries in the set SCOP-586
| Query type | Number of queries (domains) | Program | Number of assigned domains | Assignment accuracy (%) | Unassigned domain percentage (%) | Average time per query (s) | Relative to fastSCOP |
|---|---|---|---|---|---|---|---|
| Single domain | 464 query proteins (464 domains) | 3D-BLAST | 464 | 94.4% (95.9%a) | 0% | 1.166 | 0.38 |
| MAMMOTH | 464 | 98.7% (98.7%a) | 0% | 1046.47 | 338.61 | ||
| fastSCOP | 455 | 98.5% (99.6%a) | 1.94% | 3.09 | 1 | ||
| Multiple domain | 122 query proteins (272 domains) | 3D-BLAST | 275 | 86.9% | 1.8% | 2.238 | 0.34 |
| MAMMOTH | 238 | 94.1% | 12.5% | 1859.80 | 278.40 | ||
| fastSCOP without reassignmentb | 214 | 98.6% | 19.48% | 5.11 | 0.76 | ||
| fastSCOP | 254 | 98% | 6.6% | 6.68 | 1 |
aAssignment accuracy at SCOP fold level.
bfastSCOP does not apply the reassignment step, which is step 4 in Figure 1A.
SCOP-586 consists of 586 query proteins, which are in SCOP1.69 but not in SCOP1.67; the search database is SCOP1.67.
Time was measured using a personal computer with an Intel Pentium 2.8 GHz processor with 1024 MB of RAM.
Table 1 presents the accuracy of superfamily assignment and the average execution time of the fastSCOP, 3D-BLAST and MAMMOTH on the query set SCOP-586. Stand-alone fastSCOP, 3D-BLAST and MAMMOTH were run on a personal computer with a single Pentium 2.8 GHz processor with 1024 MB RAM. The 3D-BLAST and MAMMOTH used E-values and Z-scores, respectively, to order the hit proteins. For 3D-BLAST, the top rank of a hit list of a query was selected as the SCOP superfamily. For MAMMOTH, the same criteria (Z-score > 5.5; RMSD value < 4 Å and (Z-score–RMSD> 4.0) of the fastSCOP were adopted to assign a query protein to a SCOP superfamily.
On average, the fastSCOP took ∼3.09 s to recognize the structural domain and classification assignment for a single-domain query protein in the query set SCOP-586 (Table 1). It was ∼338 times faster than MAMMOTH and was ∼2.6 times slower than 3D-BLAST, because the fastSCOP required the time of applying MAMMOTH for structure alignments between the query protein and the top 10 hit domains. For multiple-domain query proteins, the fastSCOP was ∼278 times faster than MAMMOTH and was ∼2.7 times slower than 3D-BLAST. The predicted domain boundaries of the fastSCOP server were lightly shifted and the accuracy of boundaries accurate within 15 residues was 92% for the set SCOP-586.
As shown in Table 1, the fastSCOP server yielded 98.5 and 99.6% assignment accuracies at the superfamily and fold levels, respectively, for 464 single-domain queries. It outperformed 3D-BLAST (94.4 and 95.9% at the superfamily and fold levels, respectively) and performed similarly to MAMMOTH (98.7 and 98.7%). The unassignment percentage of the fastSCOP is 1.94% (nine query proteins), which slightly exceeds those of the other two methods. For 122 multiple-domain queries (with 272 domains), the fastSCOP yielded a 98.6% (214 domains) assignment accuracy and the unassignment percentage was 19.48% (53 domains) when the reassignment step (step 4 in Figure 1A) was not applied. However, the assignment accuracy was 98% (254 domains) and the unassignment percentage was reduced to 6.6% (18 domains) when the fastSCOP used the reassignment step. The accuracy of fastSCOP significantly exceeded that of MAMMOTH (94.1%) and 3D-BLAST (86.9%); the unassignment percentage was lower than that of MAMMOTH (12.5%, 34 domains).
The fastSCOP was evaluated using the 8700 PDB entries, which have no annotations in the SCOP database, and whose publishing date range from 1 January 2006 to 5 december, 2006. The fastSCOP used these 8700 protein structures as queries, and the search classification database was SCOP 1.71. In this set, 22% (1594 proteins) queries were multi-domain proteins. The fastSCOP server can automatically assign 7311 (84%) proteins (9420 domains) to the SCOP superfamilies in 9.6 h. According to the assignment accuracy (∼98%) of the fastSCOP applied to the query set SCOP-586 and the assignment criteria (step 2 in Figure 1A), the fastSCOP server accurately assigns ∼9000 domains.
CONCLUSION
This work demonstrated the robustness and feasibility of the fastSCOP server for recognizing the structural domains and the evolutionary classifications of protein structures. The key contribution of this work is the cooperative integration in fastSCOP of 3D-BLAST (a fast structural database search tool) and MAMMOTH (a fast detailed structural alignment tool); the former is required for efficiency and the latter for accuracy. Future works will adopt the fastSCOP for other evolutionary classification databases, such as CATH (4). Additionally, the fastSCOP can be applied to develop structural motifs and sequence motifs from multiple structure and sequence alignments.
ACKNOWLEDGEMENTS
J.-M.Y. was supported by National Science Council and partial support of the ATU plan by MOE. Authors are grateful to both the hardware and software supports of the Structural Bioinformatics Core Facility at National Chiao Tung University. Funding to pay the Open Access publication charges for this article was provided by National Science Council.
Confliat of interest statement. None dealared.
REFERENCES
- 1.Burley SK, Almo SC, Bonanno JB, Capel M, Chance MR, Gaasterland T, Lin D, Sali A, Studier FW. Structural genomics: beyond the human genome project. Nat. Genet. 1999;23:151–157. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- 2.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, et al. The CATH domain structure database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 6.Cheek S, Qi Y, Krishna SS, Kinch LN, Grishin NV. SCOPmap: automated assignment of protein structures to evolutionary superfamilies. BMC Bioinformatics. 2004;5:197. doi: 10.1186/1471-2105-5-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Scie. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol Crystallogr. 2002;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 11.Yang JM, Tung CH. Protein structure database search and evolutionary classification. Nucleic Acids Res. 2006;34:3646–3659. doi: 10.1093/nar/gkl395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin AC. The ups and downs of protein topology: rapid comparison of protein structure. Protein Eng. 2000;13:829–837. doi: 10.1093/protein/13.12.829. [DOI] [PubMed] [Google Scholar]
- 13.Carpentier M, Brouillet S, Pothier J. YAKUSA: a fast structural database scanning method. Proteins Struct., Funct. Genet. 2005;61:137–151. doi: 10.1002/prot.20517. [DOI] [PubMed] [Google Scholar]
- 14.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 15.Granzin J, Eckhoff A, Weiergraeber OH. Crystal structure of a multi-domain immunophilin from Arabidopsis thaliana: a paradigm for regulation of plant ABC transporters. J. Mol Biol. 2006;364:799–809. doi: 10.1016/j.jmb.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 16.Wu B, Li P, Liu Y, Lou Z, Ding Y, Shu C, Ye S, Bartlam M, Shen B, et al. 3D structure of human FK506-binding protein 52: implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc. Natl Acad. Sci. USA. 2004;101:8348–8353. doi: 10.1073/pnas.0305969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang J, Hung DT, Schreiber SL, Clardy J. Structure of the human 25 kDa FK506 binding protein complexed with rapamycin. J. Am. Chem. Soc. 1996;118:1231–1232. [Google Scholar]
- 18.Gopalan G, He Z, Balmer Y, Romano P, Gupta R, Buchanan BB, Swaminathan K, Luan S. Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen. Proc. Natl Acad. Sci. USA. 2004;101:13945–13950. doi: 10.1073/pnas.0405240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh S, Decenzo MT, Livingston DJ, Pearlman DA, Navia MA. Conformation of Fk506 in X-ray structures of its complexes with human recombinant Fkbp12 mutants. Bioorg. Med. Chem. Lett. 1995;5:1983–1988. [Google Scholar]
- 20.Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-Kalman Z, Koncz C, et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell. 2003;14:4238–4249. doi: 10.1091/mbc.E02-10-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sangster TA, Queitsch C. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 2005;8:86–92. doi: 10.1016/j.pbi.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]