Abstract
We present a new version of the European Bioinformatics Institute Web Services, a complete suite of SOAP-based web tools for structural and functional analysis, with new and improved applications. New functionality has been added to most of the services already available, and an improved version of the underlying framework has allowed us to include more applications.
Information on the EBI Web Services, tutorials and clients can be found at http://www.ebi.ac.uk/Tools/webservices.
INTRODUCTION
Web Services technology enables scientists to access EBI data and analysis applications as if they were installed on their laboratory computers. Similarly, it enables programmers to build complex applications without the need to install and maintain the databases and analysis tools and without having to take on the financial overheads that accompany these. Moreover, Web Services provide easier integration and interoperability between bioinformatics applications and the data they require.
All that is needed by the user is a lightweight program that communicates with the servers running at the EBI. These services have several advantages: they provide an easy and flexible way to deal with repetitive tasks such as bulk submission with minimal intervention from the user, and allow the programmer as well as the service provider to integrate and build more complex analysis workflows using existing EBI services.
MOTIVATION
The challenge of unravelling gene function and better understand gene regulation processes in an era where exponentially growing amounts of genomic data are being deposited into the public databases, requires fast and unlimited access to tools that can, in a systematic manner, simplify the analysis of these data.
Equally important, scientists are no longer bound to work within the confinement of their own labs. The Internet has provided the means to develop systems with which it is possible to exchange results and partial analysis of data. Characterizing a gene in terms of a sequence, its translation, expression profile, function and structure requires access to widely distributed services. The integration of such services and their interoperability is now feasible using Web Services technologies.
These data and the corresponding analysis tools are mainly accessed using browser-based interfaces. When large amounts of data need to be retrieved and analysed, this often proves to be tedious and impractical. Moreover, research is rarely completed just by retrieving or analysing a particular nucleotide or protein sequence. Database information retrieval and analysis services have to be linked, so that, for example, search results from one database can be used as the base of a search in another, the results of which are then analysed. When performing these operations using a web browser, researchers are forced to repeat the troublesome tasks of searching; copying the results for subsequent searches into other database services, and again copying the results from these for further analysis.
Creating a local bioinformatics work environment is possible by downloading and installing the necessary database content and services (such as retrieval and analysis programs). This has the advantage that processes that otherwise require manual operations can be automated. However, the hidden overheads imposed by maintaining and operating such environments are, more often than not, exceed the capacity of local systems.
Programmatic Web Services technology has gained much attention as an open architecture enabling interoperability among applications across heterogeneous platforms and different networks. The European Bioinformatics Institute (EBI) has been using this technology (1) to enhance and ease the use of the bioinformatics resources it provides (2). Currently, the European Bioinformatics Institute provides access to more than 200 databases and to about 150 bioinformatics applications.
METHODS
To ensure software from various sources work well together, this technology is built on open standards such as Simple Object Access Protocol (SOAP, http://www.w3.org/TR/soap/), a messaging protocol for transporting information; (WSDL, http://www.w3.org/TR/wsdl), a standard method of describing Web Services and their capabilities, and Universal Description, Discovery and Integration (UDDI, http://www.uddi.org/specification.html), a platform-independent, XML-based registry for services. For the transport layer itself, Web Services can use most of the commonly available network protocols, especially Hypertext Transfer Protocol (HTTP).
EBI Web Services are described by WSDL files. WSDL is an XML format for describing network services as a set of endpoints operating on messages containing either document-oriented or procedure-oriented information. The functionality provided by the service and the messages exchanged are described in an abstract manner.
A WSDL binding describes how the service is bound to a messaging protocol, particularly the SOAP messaging protocol. A WSDL SOAP binding can be either a Remote Procedure Call (RPC) style binding or a document-style binding. A SOAP binding can also have an encoded use or a literal use. We are using rpc/encoded style currently, and will be providing document/literal style soon following recommendations from the Web Services Interoperability Organization (WS-I, http://www.ws-i.org) guidelines. For a detailed explanation of the differences between both styles, see http://www-128.ibm.com/developerworks/webservices/library/ws-whichwsdl/.
A client (program) connecting to a Web Service can read the WSDL to determine what functions are available on the server. Any special data types used are embedded in the WSDL file in the form of an XML Schema. The client can then use SOAP to actually call one of the functions listed in the WSDL. Most SOAP frameworks and toolkits provide methods for the automatic generation of client code from the WSDL description.
SERVICES DESCRIPTION
Currently, EBI supports SOAP services for both database information retrieval and sequence analysis. Information about these services can be accessed from the web page http://www.ebi.ac.uk/Tools/webservices (Table 1).
Table 1.
Web Services available at the European Bioinformatics Institute
| Application | Web Services |
|---|---|
| Data retrieval | WSDbfetch, ChEBI WS, Integr8 WS, MSD API, Citexplore, OntologyLookup, Martservice |
| Analysis tools | InterProScan, Emboss |
| Homology searches | Fasta, WU-Blast, NCBI Blast, PSI-Blast, MPsrch, ScanPS |
| Multiple sequence alignment | ClustalW, KAlign, Mafft, Muscle, T-Coffee |
| Structural analysis | DaliLite, Maxsprout, SSM |
| Text mining | Whatizit |
Data retrieval
WSDbfetch allows retrieving entries in various common formats from more than 20 biological databases including EMBL (3), UniprotKB (4), Interpro (5), etc. It provides several methods for retrieving information about the service (getAvailableDatabases, getAvailableFormats, getAvailableStyles) and a fetchData operation for the actual retrieval. The user just needs to provide the database name and database identifier or accession number, and retrieve the entry (or entries) in either ASCII text, HTML with hyperlinks or XML. An example of a simple Ruby (http://www.ruby-lang.org/en/) client is presented subsequently (Figure 1).
Figure 1.
Example of a Ruby client for WSDbfetch.
Similarity search tools
A first step in many analysis procedures is usually to carry out a primary database search in order to identify sequence similarities and several algorithms are available to compare nucleotide or protein queries with nucleotide or protein databases.
Basic local alignment search tool (BLAST) (6) is probably the most popular sequence similarity search program. The EBI provides NCBI BLAST (including PHI-BLAST and PSI-BLAST (7)) and WU-BLAST (http://blast.wustl.edu) servers with a common homepage at http://www.ebi.ac.uk/blast/and a FASTA (8) server at http://www.ebi.ac.uk/fasta/. Figure 2 shows an example of Ruby client for WU-Blast.
Figure 2.
Example of a Ruby client for WU-Blast.
Apart from Blast and Fasta, EBI provides two protein-specific search tools. MPsrch (http://www.ebi.ac.uk/MPsrch/) is a biological sequence sequence comparison tool that implements the true Smith and Waterman algorithm (9). It allows a rigorous search in a reasonable computational time. SCANPS (Scan Protein Sequence, http://www.ebi.ac.uk/scanps/) is another program for comparing a protein sequence to a database of protein sequences. It also implements the full Smith–Waterman style searching and is capable of identifying multiple domain matches by using iterative profile searching. Both methods are available in our Web Services suite.
InterProScan
InterPro is an integrated resource for protein families, domains and functional sites, which integrates the following protein signature databases: PROSITE, PRINTS, ProDom, Pfam, SMART, TIGRFAMs, PIRSF, SUPERFAMILY, Gene3D and PANTHER (5).
InterProScan (10) is a tool that integrates the search algorithms and protein signature recognition methods from the InterPro member databases into one resource, and provides the corresponding InterPro accession numbers and Gene Ontology (GO) (11) annotation in the results. These GO mappings provide annotation for 61% of UniProtKB proteins, which facilitates GO annotation to query proteins. The current release of InterPro contains more than 13 000 entries, with its signatures covering over 78% of UniProtKB proteins. Figure 3 contains an example Perl (www.perl.org) client for InterProScan.
Figure 3.
Example of a Perl client for InterProScan.
Multiple and pairwise sequence alignment applications
Over the past years, multiple sequence alignments (MSAs) have become one of the most widely used tools in biology along with database search methods. MSAs are needed for profile analysis, phylogenetic reconstruction, structure prediction and a wealth of minor but important applications such as PCR primer design or sequence reconciliation. The ever-growing reliance on MSAs is even more pronounced now that hundreds of complete genomes are being made available.
CLUSTALW (12) is a widely used program for multiple sequence alignment and is one of the most popular tools at the EBI. T-Coffee (13), MUSCLE (14), MAFFT (15) and Kalign (16), are other tools that employ newer algorithms that complement the accuracy of CLUSTALW.
For pairwise alignment, dynamic programming methods ensure an optimal solution by exploring all possible alignments and choosing the best one. The European Molecular Biology Open Software Suite (EMBOSS) (17), includes the programs ‘water’, a tool implementing the Smith–Waterman for local alignments, and ‘needle’, an implementation of the Needleman-Wunsch (18) algorithm for global alignments.
All these methods are now available as Web Services from the EBI, providing a sensible framework for multiple sequence alignment processing.
Structural analysis
Tools available as Web Services for structural analysis include MaxSprout (19), which is a fast database algorithm for generating protein backbone and side chain co-ordinates from a C(alpha) trace. The backbone is assembled from fragments taken from known structures. Side chain conformations are optimized in rotamer space using a rough potential energy function to avoid clashes. Also, DaliLite (20), which, computes optimal and suboptimal protein structural alignments between two input sets of atomic coordinates.
Text mining
Whatizit (21) is a text processing system that allows you to do text mining tasks on text. The tasks are defined by a series of pipelines. The description of each text processing step can be found in the on line documentation of the tool (http://www.ebi.ac.uk/webservices/whatizit/info.jsf). Optionally, instead of providing the text to be analysed, users can supply a term query. This will result in the retrieval of publicly available (i.e in MEDLINE) abstracts matching the terms in the query and their consequent annotation by the pipeline of your choice.
Whatizit can identify molecular biology terms and link them to publicly available databases. Terms identified by the system are wrapped with XML tags that carry additional information, such as the primary keys to the databases where all the relevant information is kept. This service is highly appreciated by people who are reading literature and need to quickly find more information about the query term, e.g. its Uniprot id, MEDLINE references, UniProt/Swiss-Prot keywords, Gene Ontology (GO) terms and the NCBI Taxonomy.
The Whatizit Web Service is available as a SOAP implementation and as a streamed servlet. The methods available through the SOAP interface are presented in Table 2.
Table 2.
Methods available in the Whatizit Web Service
| getPipelineStatus | Returns a list of available pipelines (text processing tasks) together with their current status (available/under maintenance) and a description of what they do. |
| contact | Takes two parameters, the name of a pipeline and the text to be annotated. The text is sent to the specified pipeline and returned with all the annotation in place. |
| queryPmid | Takes two parameters, the name of a pipeline and a pmid. The system retrieves the specified pmid and sends it for annotation to the given pipeline. |
| search | Takes two parameters, the name of a pipeline and a term query. The system preforms the retrieval based on the term query and sends the results for annotation to the given pipeline. |
CURRENT IMPLEMENTATION
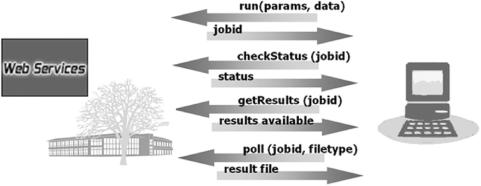
Most of the EBI services presented here are implemented using a common Perl-based framework. These are tightly integrated with EBI hardware and middleware infrastructure and provide a uniform interface to the user. SOAP::Lite 0.60 was selected as the SOAP toolkit as it has proven to be the most stable. Sun's JAX-WS RI 2.0 is used for the WSDbfetch and Whatizit implementations. These provide for basic methods: runApp, checkStatus, getResults and poll, which are summarized as follows:
The runApp method (where App is the name of the application, i.e. runFasta, runClustalW etc) is used to submit a job to the EBI job dispatcher. This method accepts two inputs: an InputParams structure with the options to be passed to the application, and a string array with the sequences. The job can be submitted in two modes: synchronous and asynchronous. In both cases, the server returns a job identifier which can be used to retrieve the results (Figure 4). Examples of client programs were shown in Figures 1–3.
Figure 4.
Methods and message flow diagram for EBI Web Services.
COMBINING WEB SERVICES
One of the main advantages of Web Services is that researchers can easily construct bioinformatics workflows and pipelines combining two or more Web Services to solve complex biological tasks such as protein function prediction, genome annotation, microarray analysis, etc. Users can customize any analytical protocol by combining services available from different locations. Services, thus become building blocks that can be exchanged, allowing flexibility and robustness. Workflow protocols can be created as either simple scripts or using graphical workflow tools such as Taverna (22) or Triana (23).
A summary of other Web Services available from the EBI is presented in Table 3. There is also a wide range of Web Services available worldwide, with those provided by the NCBI Entrez Programming Utilities (24), the DNA Databank of Japan (DDBJ) (25) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (26) being the most commonly used in bioinformatics. Additional tools and databases include PathPort/ToolBus tools (27), BioMOBY (28) BIND (29). A more comprehensive list and description of the services available can be found at the Web Services EBI pages (http://www.ebi.ac.uk/Tools/webservices/).
Table 3.
Examples of SOAP Web Services provided by the European Bioinformatics Institute
| Service | Description |
|---|---|
| CiteXplore | Retrieve data from the Citation database |
| MSD API | Access to data and tools from the Macromolecular Structures database. (1) |
| Integr8 | Provides access to a subset of the data available in Integr8 (30). |
| ChEBI | Allows you to retrieve entries from the ChEBI database. (31) |
| Martservice | Provides access to the resources in the Biomart Central Server (32) |
| OntologyLookup | Provides a Web Service interface to query multiple ontologies from a single location with a unified output format. (33) |
| SOAPLAB | Unified access to different bioinformatics packages (1) |
USAGE
More than 1 600 000 job submissions were processed using Web Services during 2006, accounting for around 30% of all jobs run at the EBI during that period. InterProScan (1 500 000+) and Blast (120 000+) are the most used services. Additionally, more than two million entries were retrieved through WSDbfetch, which accounts for 35% of all dbfetch type requests. Users involved in high-throughput processing and requiring systematic usage of a particular tool are the main beneficiaries of these services.
Commercial as well as academic bioinformatics service providers, such as ProFunc (34), ELM Server (35), the Uniprot Unified Website, Integr8 or Blast2GO (36) have adopted our services as an integral part of their online services. They are also used by many Open Source projects and commercial tools such as Jalview (37) and BlastStation (http://www.blaststation.com).
FUTURE PLANS
After a careful evaluation of existing technologies, and taking into consideration our users’ feedback, we are planning for continuous improvement and re-engineering of implementation of future services. We have chosen JAX-WS (http://java.sun.com/webservices/jaxws/) as a basis for our future Web Services infrastructure. We are confident that this change will allow us to meet the increasing demand and improve the level of service. The JAX-WS technology has reached a sufficient level of maturity and commitment by the developer and user communities, and is architectured for high performance, extension and interoperability. New features, such as WS-Security and WS-RM, (http://en.wikipedia.org/wiki/List_of_Web_service_specifications) will be introduced, ensuring that we will be able to provide advanced functionality and meet future requirements.
We will be also moving to a document/literal style WSDL descriptions following the Web Services Interoperability Organization (WS-I) guidelines, and will implement REST style (38) interfaces to most of the services. REST stands for Representational State Transfer, this basically means that each unique URL is a representation of some object. It is possible to get the contents of that object using an HTTP GET and use an HTTP POST to modify the object. It provides improved response times and server loading characteristics due to support for caching and the fact that no XML parsing is involved. Clients are easy to build, and no toolkits are required. However, tooling and infrastructure for SOAP provide greater productivity, making it a more strategic investment for a wider range of long-term requirements. More information can be obtained at http://www.ebi.ac.uk/Tools/webservices/about/rest.
CONCLUSION
We have presented here a set of services that give the user more direct access to data and services from the EBI. Users can access all data and applications as if they were installed in their local machines, providing seamless integration between disparate services and allowing the construction of workflows to perform complex tasks.
ACKNOWLEDGEMENTS
The EMBL-EBI's Web Services are supported by the European Union (contract number 021902 as part of the FELICS Research Infrastructure; contract number LHSG-CT-2004-12092 as part of the EMBRACE project; and contract number IST-2001-32688 as part of the ORIEL Project), the Wellcome Trust; the European Patent Office; the National Institutes of Heath (as part of the UniProt project, grant 1 U01 HG02712-01); and core funding from the European Molecular Biology Laboratory (EMBL). Funding to pay the Open Access publication charges for this article was provided by the EMBL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pillai S, Silventoinen V, Kallio K, Senger M, Sobhany S, Tate J, Velankar S, Golovin A, Henrick K, et al. SOAP-based services provided by the European Bioinformatics Institute. Nucleic Acids Res. 2005;33:25–28. doi: 10.1093/nar/gki491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harte N, Silventoinen V, Quevillon E, Robinson S, Kallio K, Fustero X, Patel P, Jokinen P, Lopez R. Public web-based services from the European Bioinformatics Institute. Nucleic Acids Res. 2004;32:3–9. doi: 10.1093/nar/gkh405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulikova T, Akhtar R, Aldebert P, Althorpe N, Andersson M, Baldwin A, Bates K, Bhattacharyya S, Bower L, et al. EMBL nucleotide sequence database in 2006. Nucleic Acids Res. 2007;35:16–20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35:193–197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Buillard V, Cerutti L, et al. New developments in the InterPro database. Nucleic Acids Res. 2007;35:224–228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul SF, Warren G, Webb M, Eugene WM, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. PNAS. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 10.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 14.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lassmann T, Sonnhammer E. Kalign – an accurate and fast multiple sequence alignment algorithm. BMC Bioinform. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Gen. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 18.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 19.Holm L, Sander C. Maxsprout. J. Mol. Biol. 1991;218:183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 20.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 21.Rebholz-Schuhmann D, Kirsch H. In Proceedings HDL 2004. UK: Bath; 2004. Extraction of biomedical facts - a modular Web server at the EBI (Whatizit) [Google Scholar]
- 22.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock M, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:729–732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majithia S, Shields MS, Taylor IJ, Wang I. In Proceedings of the IEEE International Conference on Web Services (ICWS’04) 2004. Triana: A Graphical Web Service Composition and Execution Toolkit; pp. 514–524. San Diego, California, USA. [Google Scholar]
- 24.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:173–180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazaki S, Sugawara H, Ikeo K, Gojobori T, Tateno Y. DDBJ in the stream of various biological data. Nucleic Acids Res. 2004;32:31–34. doi: 10.1093/nar/gkh127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in Kegg. Nucleic Acids Res. 2006;34:354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckart JD, Sobral BW. A life scientist's gateway to distributed data management and computing: the pathport/toolbus framework OMICS. 2003;7:79–88. doi: 10.1089/153623103322006661. [DOI] [PubMed] [Google Scholar]
- 28.Wilkinson M, Schoof H, Ernst R, Haase D. BioMOBY successfully integrates distributed heterogeneous bioinformatics web services. The PlaNet Exemplar Case Plant Physiol. 2005;138:5–17. doi: 10.1104/pp.104.059170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bader GD, Betel D, Hogue CW. Bind: the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:297–302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooksbank C, Cameron G, Thornton J. The European Bioinformatics Institute's data resources: towards systems biology. Nucleic Acids Res. 2005;33:D46–D53. doi: 10.1093/nar/gki026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, et al. EnsMart: A generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cote RG, Jones P, Apweiler R, Hermjakob H. The ontology lookup service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinform. 2006;28;7:97. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puntervoll P, Linding R, Gemünd C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DMA, Ausiello G, Brannetti B, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conesa A, Goetz S, Garcia JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 37.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java Alignment Editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 38.Roy T. PhD thesis. 2000. Fielding, ‘Architectural Styles and the Design of Network-based Software Architectures’. UC Irvine. [Google Scholar]