Abstract
We present the ISACGH, a web-based system that allows for the combination of genomic data with gene expression values and provides different options for functional profiling of the regions found. Several visualization options offer a convenient representation of the results. Different efficient methods for accurate estimation of genomic copy number from array-CGH hybridization data have been included in the program. Moreover, the connection to the gene expression analysis package GEPAS allows the use of different facilities for data pre-processing and analysis. A DAS server allows exporting the results to the Ensembl viewer where contextual genomic information can be obtained. The program is freely available at: http://isacgh.bioinfo.cipf.es or within http://www.gepas.org.
INTRODUCTION
Genetic aberrations, such as losses (deletions) or gains (amplifications) of genetic material that affect certain regions of the genome, have been shown to be on the basis of many human pathologies, including rare diseases, as mental retardation (1), or much more prevalent pathologies, as cancer (2).
Classical approaches to characterize these genetic aberrations used comparative genomic hybridization (CGH), in which genomic DNA was hybridized to metaphase chromosomes (3). Recently, however, the use of different types of microarrays to directly study genomic variations in DNA copy number is becoming more and more popular. Such massive genomic approaches are known as array comparative genomic hybridization, or Array CGH (4). Different options are used to implement Array CGHs including large genomic clones (5), cDNAs (6), oligonucleotides (7) and even SNP genotyping platforms (8). These new technologies along with the use of expression arrays offer for the first time the opportunity of characterize in an accurate way the dependence of gene expression on alterations in genomic copy number (9,10).
As in other high-throughput methodologies, data analysis and, in particular, biological interpretation of the results constitutes a well-known bottleneck. Specific problems related to the analysis of Array CGH can be circumscribed mainly to: (i) the accurate definition of the borders of the genetic alteration and the copy number estimation, (ii) the appropriate mapping and visualization of the data onto the chromosomes and (iii) the possibility of formulating reasonable hypothesis that link genes to diseases by understanding the alteration of the functions at molecular level. The first aspect has been the motivation for a number of analytical approaches recently proposed (11,12). Although several programs have been developed for array-CGH data visualization and analysis, almost all of them are stand-alone applications in different programming languages such as R and MATLAB scripts, C or java (12). To our knowledge only two web-based applications for array-CGH data analysis have been published to date: CAPweb (13) and ArrayCyGHt (14). Among the specific problems previously mentioned, probably, the last one is the most relevant given that the ultimate aim of studies of copy number chromosomal alterations is to understand what is the functional effect produced at molecular level that can help to interpret the pathologic phenotype. In the classical vision, one or a few key genes are the causative factors for this type of pathologies, and the problem consisted in identifying such genes within the region amplified or deleted. This vision is changing by the recent report of regions in the chromosomes of higher eukaryotes containing coexpressing genes (15) which, in addition, are functionally related (16). Actually, regional arrangements of genes have found to be regulated not only by copy number alterations but also by different mechanisms such as epigenetic modifications (17). This reinforces the functional role of chromosomal regions containing groups of functionally related genes and their possible impact on diseases such as cancer (18). This important aspect, however, remains mostly overlooked in the tools for the analysis of copy number alterations.
We present here the ISACGH program that allows visualizing array CGH data or/and expression arrays onto human or mouse chromosomal coordinates (automatically found through their standard identifiers) and represents the regions with copy number alterations found by using different methods. Correlations between copy number and gene expression level can be visualized in different plots. The program finds minimal common regions with altered copy number across different arrays. Although ISACGH can be used alone, it is tightly integrated into the GEPAS (19,20) and Babelomics (21) packages. Thus, normalization and any other data transformation operations can directly be performed within a common environment, without the necessity of reformatting the data. The connection of ISACGH to different tools for functional profiling (21,22) offer the possibility of studying the enrichment in functionally relevant terms (gene ontology, pathways, etc) in chromosomal regions with copy number alterations.
FUNCTIONALITY AND VISUALIZATION
The program
ISACGH (a meta acronym that stands for In Silico Array-CGH) is a web-based integral system that allows studying, within the same context, copy number alterations and gene expression, and provides facilities for the functional profiling of the regions affected. ISACGH can process most of the common gene identifiers and automatically maps them onto chromosome coordinates (human or mouse are available). ISACGH can input gene expression values, genomic hybridization values or both simultaneously. It is not necessarily to use the same platform for chromosomal and expression hybridizations. For example, a case in which a BAC array is used for copy number analysis and a cDNA array is used for gene expression analysis can be analyzed. In principle the number of probes that can be handled depends mainly on the browser used and the memory of the client computer. Current browsers can easily handle high density arrays in the order of 100 000 probes or even more.
Input format
The input format is the one used by GEPAS (19,20) and other similar tools and consists of a tab-delimited text file where the first column correspond to the probe identifiers. The following column(s) correspond to the hybridization intensities (or ratios if two-colour microarrays are used) obtained for each probe in the microarray(s) analyzed. Either genomic hybridizations or mRNA-derived hybridizations are input in the same format. Additionally a file with the chromosomal coordinates of the probes in the chromosomes can be provided. Again, this is a tab-delimited text file with four columns: the fist one contains the probe identifiers, the second one the chromosome in which these are located and the third and fourth ones the chromosome coordinates of the 5′ and 3′ ends of the probes.
Functionality and representation of the results
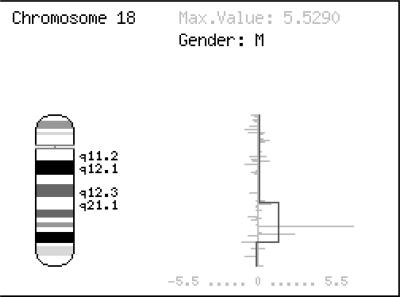
When genomic hybridization is used, the program predicts the regions with copy number alterations. If only gene expression values are provided, these are mapped onto their chromosomal coordinates. When both, genomic and gene expression values are provided, changes in genomic copy number are predicted and plotted in the same figure together with expression values. Figure 1 shows a combined plot of copy number estimation (blue line) and gene expression (grey bars) in the human chromosome 18. An important aspect is the assessment of the effect of copy number in the global expression of the genes contained in the amplified/lost region. To this end a Student t-test has been implemented to assess differential expression between the genes with normal copy number (those in the base line block) and the genes found in regions with copy number alterations. In addition, plots for the direct visualization of the relationship between both expression and copy number can be obtained. Interestingly, if expression values are entered instead of genomic hybridization values, the program can find regions of increased gene expression (RIDGEs) (15).
Figure 1.
Human chromosome 18. Multiple myeloma (mm) cell line SK-MM-2 (see text) with copy number estimation (blue line) and gene expression values (grey bars). The isowindow segmentation method was used to estimate significant alterations in copy number.
There are different possibilities for the representation of the results which include several types of multiple-view plots (all the chromosomes of one sample or the same chromosome for multiple samples). In addition, plots of piled samples to detect minimal regions with deletions (or amplifications) in the chromosomes can be obtained. All the results obtained can be visualized in detail in the ISACGH internal viewer but, as an additional and novel feature, they can also be visualized onto the Ensembl browser.
The distributed annotation system (DAS) is a client-server system in which a single client, in this case the Ensembl (http://www.ensembl.org), integrates information from multiple servers (see http://www.biodas.org). Using the DAS architecture, the Ensembl gathers genome annotation information from multiple distant web sites, collate such information, and display it to the user in its viewer together with the own ensemble data and predictions. Thus, the use of DAS servers for visualization of any genomic feature on the Ensembl viewer offer an excellent environment for the study of the results produced by ISACGH in the genomic context, with the possibility of accessing to any type of available information.
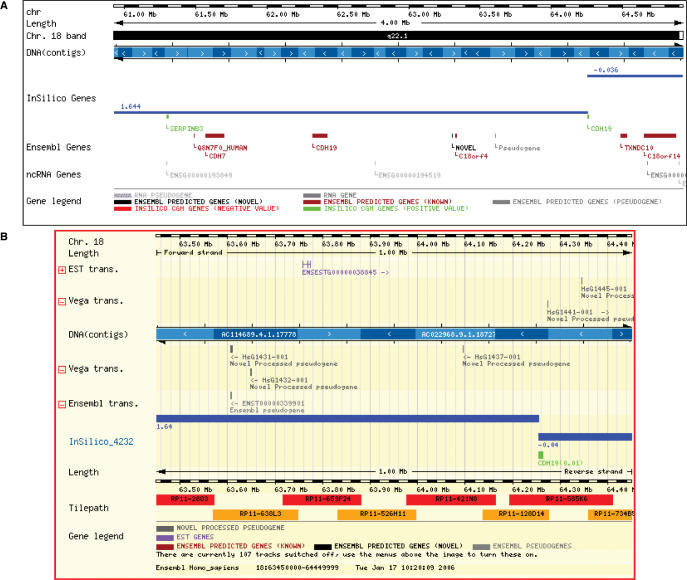
Then, if the Ensembl DAS server option is selected, clicking onto a chromosomal region will produce the creation of a DAS server with information about the probes in the region and the copy number estimation. This information is exported to the Ensembl viewer, which acts as DAS client. Figure 2B shows approximately the same chromosomal region than Figure 2A, but represented in the Ensembl environment. Any genomic feature available in Ensembl in the same chromosomal region can be visualized together with the ISACGH results.
Figure 2.
The two zoom options in the breakpoint on the extreme closest to the centromer of the amplicon detected in 18q21.1 in one of the mm cases studied. The two probes form the array shown in the figure (the ones corresponding to SERPINB3 and CDH19) are green because all of them represent amplifications. The blue line represents the copy number estimation. (A) ISACGH viewer, (B) DAS server.
Breakpoint detection
Two methods for breakpoint detection, GLAD (23) and CBS (24), which are among the best performers (11) have been included in the program. We have also developed and included two new methods: a segmentation method (isowindow) and a method based on the slopes of regression in local intervals for copy number change detection. A comparison of the relative performances of the methods implemented was carried out by means of simulated data sets. The new methods proposed here perform at least as well as the GLAD and CBS in terms of tolerance to noise and accuracy in the determination of breakpoints but are more efficient in terms of runtimes (data available in http://bioinfo.cipf.es/downloads/).
Functional profiling of regions with copy number alterations
As previously commented, the ultimate aim of an Array-CGH experiment is to find a molecular explanation for the effects of the detected copy number alterations. The interpretation of genome-scale data is usually performed in two steps: in a first step, genes of interest are selected in this case because they are located in the amplified (or lost) region detected. In a second step, the selected genes of interest are compared to the background (here the rest of genes in the chromosome) in order to find enrichment in any functional category (gene ontology, KEGG pathways, etc.) This comparison to the background is required because otherwise the significance of a proportion (even if high) cannot be determined. Different approaches have been developed to this end (25). Here we will use the FatiGO (22) method, which uses a Fisher's exact test to determine the enrichment in different functional categories. In this case we will analyse the enrichment in GO terms but other functional categories such as KEGG pathways, Interpro functional motifs, Swissprot keywords and some regulatory elements as transcription factor binding sites or other regulatory motifs can also be analyzed with this tool.
A CASE STUDY OF MULTIPLE MYELOMA
To illustrate the concept of functional profiling in the context of array CGH we will use an example of multiple myeloma (MM), an incurable form of haematological neoplasia. The data and the experimental steps followed are described in (26). The aim here was to identify any possible region that contained copy number gains (amplifications), to study the expression of the genes included in that particular region and to understand the possible functional consequences of such alterations.
Data from two-colour hybridizations for both nuclear DNA and transcripts were normalized using the corresponding GEPAS (19,20) module DNMAD and redirected to ISACGH from there. The isowindow method, at medium resolution, was used as the option for the estimation of regions with copy number alterations. The aim was to identify the amplified regions (amplicons) and, to localize and identify the genes that are placed at the amplicon limits. The next step involved the determination of the global expression status of the genes included in these amplicons. And the final aim was to understand the functional consequences associated to the alteration of the expression of such genes.
The analysis was focussed in the chromosome 18, where high level amplification and recurrent gains were found by conventional CGH in cell lines or primary patient samples (27). Within this chromosome, a region with a high level of amplification (amplicon) located at the cytoband 18q21 was detected. MM cell line SK-MM-2 showed a well defined amplicon with an altered gene expression profile (Figure 1). Within the limits of the amplified region several genes display higher expression rates (Figure 1).
Functional profiling of the amplicon revealed a significant enrichment in a number of GO terms in the genes contained in such region. Thus, the GO terms regulation of cellular process (GO:0050794) and regulation of physiological process (GO:0050791) were significantly over-represented in the amplicon (FDR adjusted p-value = 0.0336). Genes annotated with these terms were: BCL2, MALT1, NEDD4L, MBD2, TNFRSF11A and TCF4. Some of them have annotations at more detailed levels in GO, although the number of genes is too small as to produce statistically significant results. For example BCL2 and MALT1 are annotated as negative regulation of programmed cell death (GO:0043069). These results show how the amplification is affecting to a group of functionally related genes and allows conjecturing their global implication in the diseased condition.
DISCUSSION
We present ISACGH, a web-based integrated system that allows simultaneously studying copy number alterations using array-CGH, their effect on gene expression and the possible functional impact of the chromosomal alteration. In addition, ISACGH is integrated in the GEPAS package, facilitating the normalization, data transformation and other higher-level analysis such as differential gene expression, clustering, etc. This integration may help researchers to overcome the necessity of cumbersome data reformatting operations. Although other two web-based applications for array-CGH data analysis are available [CAPweb (13) and ArrayCyGHt (14)], ISACGH is the only web-based tool offering this combination of analyses to our knowledge.
The results obtained in the case study suggest that the alterations that ultimately lead to MM are not produced by the deregulation of one unique gene, but are rather the combined result of simultaneous deregulations of genes involved in one or more pathways or biological functions. Recent observations on the existence of a non-negligible number of clusters of functionally- related genes suggests that this phenomenon might be more frequent in pathologies characterized by copy number alterations than previously imagined. These findings stress on the importance of the functional profiling for the proper understanding of the functional implications of genomic copy number alterations.
ACKNOWLEDGEMENTS
This work is supported by grants from the Spanish ministry of education and science (BIO 2005-01078) and National Institute of Bioinformatics (www.inab.org) a platform of Genoma España. Funding to pay the Open Access publication charges for this article was provided by Genoma España.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bassett AS, Chow EW, Weksberg R. Chromosomal abnormalities and schizophrenia. Am. J. Med. Genet. 2000;97:45–51. doi: 10.1002/(sici)1096-8628(200021)97:1<45::aid-ajmg6>3.0.co;2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertson DG, Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum. Mol. Genet. 2003;12:R145–R152. doi: 10.1093/hmg/ddg261. [DOI] [PubMed] [Google Scholar]
- 3.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 4.Mantripragada KK, Buckley PG, de Stahl TD, Dumanski JP. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat. Genet. 2005;37(Suppl):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 6.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho B, Ouwerkerk E, Meijer GA, Ylstra B. High resolution microarray comparative genomic hybridization analysis using spotted oligonucleotides. J. Clin. Pathol. 2004;57:644–646. doi: 10.1136/jcp.2003.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X, Mok SC, Chen Z, Li Y, Wong DT. Concurrent analysis of loss of heterozygosity (LOH) and copy number abnormality (CNA) for oral premalignancy progression using the Affymetrix 10K SNP mapping array. Hum. Genet. 2004;115:327–330. doi: 10.1007/s00439-004-1163-1. [DOI] [PubMed] [Google Scholar]
- 9.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- 10.Mahlamaki EH, Kauraniemi P, Monni O, Wolf M, Hautaniemi S, Kallioniemi A. High-resolution genomic and expression profiling reveals 105 putative amplification target genes in pancreatic cancer. Neoplasia. 2004;6:432–439. doi: 10.1593/neo.04130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai WR, Johnson MD, Kucherlapati R, Park PJ. Comparative analysis of algorithms for identifying amplifications and deletions in array CGH data. Bioinformatics. 2005;21:3763–3770. doi: 10.1093/bioinformatics/bti611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lockwood WW, Chari R, Chi B, Lam WL. Recent advances in array comparative genomic hybridization technologies and their applications in human genetics. Eur. J. Hum. Genet. 2006;14:139–148. doi: 10.1038/sj.ejhg.5201531. [DOI] [PubMed] [Google Scholar]
- 13.Liva S, Hupe P, Neuvial P, Brito I, Viara E, La Rosa P, Barillot E. CAPweb: a bioinformatics CGH array analysis platform. Nucleic Acids Res. 2006;34:W477–W481. doi: 10.1093/nar/gkl215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SY, Nam SW, Lee SH, Park WS, Yoo NJ, Lee JY, Chung YJ. ArrayCyGHt: a web application for analysis and visualization of array-CGH data. Bioinformatics. 2005;21:2554–2555. doi: 10.1093/bioinformatics/bti357. [DOI] [PubMed] [Google Scholar]
- 15.Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 16.Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 17.Stransky N, Vallot C, Reyal F, Bernard-Pierrot I, de Medina SG, Segraves R, de Rycke Y, Elvin P, Cassidy A, et al. Regional copy number-independent deregulation of transcription in cancer. Nat. Genet. 2006;38:1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Luoh SM, Zhang Y, Watanabe C, Wu TD, Ostland M, Wood WI, Zhang Z. Genome-wide identification of chromosomal regions of increased tumor expression by transcriptome analysis. Cancer Res. 2003;63:5781–5784. [PubMed] [Google Scholar]
- 19.Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montaner D, Tarraga J, Huerta-Cepas J, Burguet J, Vaquerizas JM, Conde L, Minguez P, Vera J, Mukherjee S, et al. Next station in microarray data analysis: GEPAS. Nucleic Acids Res. 2006;34:W486–W491. doi: 10.1093/nar/gkl197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. BABELOMICS: a suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 23.Hupe P, Stransky N, Thiery JP, Radvanyi F, Barillot E. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20:3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- 24.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 25.Dopazo J. Functional interpretation of microarray experiments. Omics. 2006;10:398–410. doi: 10.1089/omi.2006.10.398. [DOI] [PubMed] [Google Scholar]
- 26.Largo C, Alvarez S, Saez B, Blesa D, Martin-Subero JI, Gonzalez-Garcia I, Brieva JA, Dopazo J, Siebert R, et al. Identification of overexpressed genes in frequently gained/amplified chromosome regions in multiple myeloma. Haematologica. 2006;91:184–191. [PubMed] [Google Scholar]
- 27.Cigudosa JC, Rao PH, Calasanz MJ, Odero MD, Michaeli J, Jhanwar SC, Chaganti RS. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998;91:3007–3010. [PubMed] [Google Scholar]