Abstract
g:Profiler (http://biit.cs.ut.ee/gprofiler/) is a public web server for characterising and manipulating gene lists resulting from mining high-throughput genomic data. g:Profiler has a simple, user-friendly web interface with powerful visualisation for capturing Gene Ontology (GO), pathway, or transcription factor binding site enrichments down to individual gene levels. Besides standard multiple testing corrections, a new improved method for estimating the true effect of multiple testing over complex structures like GO has been introduced. Interpreting ranked gene lists is supported from the same interface with very efficient algorithms. Such ordered lists may arise when studying the most significantly affected genes from high-throughput data or genes co-expressed with the query gene. Other important aspects of practical data analysis are supported by modules tightly integrated with g:Profiler. These are: g:Convert for converting between different database identifiers; g:Orth for finding orthologous genes from other species; and g:Sorter for searching a large body of public gene expression data for co-expression. g:Profiler supports 31 different species, and underlying data is updated regularly from sources like the Ensembl database. Bioinformatics communities wishing to integrate with g:Profiler can use alternative simple textual outputs.
INTRODUCTION
High-throughput technologies have paved the road to a new era in molecular biology, allowing us to study the behaviour and relationships of many genes and molecules in parallel. With new experimental techniques, one can perform high-throughput measurements of mRNA and protein expression levels, DNA methylation status, molecule interactions, genotypes and other polymorphisms (1–4).
A critical challenge is to bring order and understanding into this exponentially growing amount of data. Easy-to-use terminology in conjunction with computational and statistical methods is needed to convert the low-level noisy data into high-level biological understanding. The Gene Ontology Consortium (http://www.geneontology.org) has been a great success in developing precise terminology that can be used across many different species. GO provides structured means to describe the biological processes, cell components and molecular functions of gene products (5). Millions of genes and gene products from many species have been annotated to the categories of GO using the best currently available knowledge (6).
Equipped with such information, one can develop methods that automatically infer knowledge about the common characteristics of genes that have been identified in high-throughput methods. This was already envisaged in the early days of GO development and is found to be very true by looking at the tools listings on the GO Consortium web site. A considerable amount of software for ontological analysis of gene lists has been published over the last 5 years, each of them having advantages and drawbacks and each approaching data or vocabularies in a slightly different manner. Babelomics (7), Onto-Tools (8) and DAVID (9) are among some of the most well-known tools. A recent review (10) gives insight to the variety of available tools and points out some open questions and drawbacks common to many or all of the compared tools.
With g:Profiler, we address several challenges that still exist in making such analysis more useful for typical end users. The distinguishing features of g:Profiler are the ease of use and informative visual presentation of the results, both well appreciated by biologists. To verify the statistical significance of the results, we have developed a new method for estimating the effect of multiple testing over large and complex structures like GO. An important feature of g:Profiler is the support for ranked gene lists that are analysed by finding functional enrichments from the top of the list. Other features that simplify the life of users include the support of most gene identifier types that can be used even mixed, conversions between different ID types, ability to find orthologous genes from other species and search for co-expressed genes based on public microarray data. The unique feature of g:Profiler compared to other GO mining tools is the integration of several functional profiling resources into a single web tool that provides a unified view to functional enrichment of unordered and ordered gene lists, mapping IDs between different types, mapping IDs to their orthologs and searching across many public gene expression data sets.
g:Profiler Web Toolset
g:Profiler is a freely available collection of web tools dedicated to the analysis of high-throughput data. It consists of four well-integrated modules: 1) g:Profiler core for functional profiling of flat or ranked gene lists; 2) g:Convert for gene identifier conversions; 3) g:Orth for fetching orthologous genes; and 4) g:Sorter for searching co-expressed genes from public data. All tools are highly interactive and cross-linked to allow rapid and fruitful analysis of versatile data. Next, we describe these modules in more detail.
g:PROFILER core—functional characterisation of flat or ranked lists of genes
Primary input to g:Profiler is a list of gene, protein, or probe identifiers from any of the currently supported 31 species. g:Profiler supports many ID types and even mixing of arbitrary ID types. This is important for a user, and it also simplifies cross-linking from other tools and web sites.
Typical sources of such a list may, for instance, be a cluster of co-expressed genes from a microarray experiment (1,11), predicted network of interacting proteins (3), direct targets of transcription factors (TFs) from genome-wide localisation studies (4), or genes with predicted TF-binding sites in their regulatory regions (12). The purpose of g:Profiler is to find common high-level knowledge such as pathways, biological processes, molecular functions, subcellular localisations, or shared TF-binding sites (TFBS) to the list of input genes. The data used in g:Profiler is derived from the Gene Ontology (5), KEGG (13), Reactome (14) and TRANSFAC (15) databases.
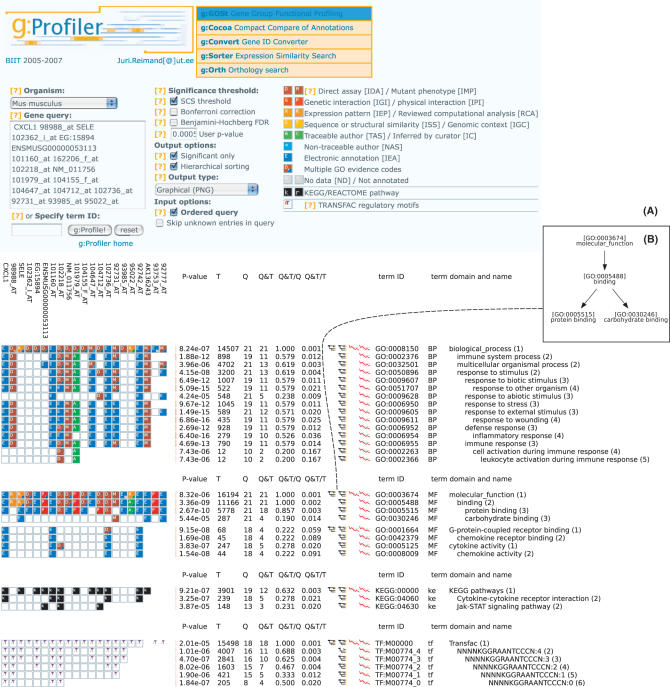
The typical result of g:Profiler analysis is a set of enriched functional terms from GO and other relevant biological databases. The resulting terms are presented in either tree-like top-down order grouped by domains, or ranked by statistical significance. Every term is accompanied by the size of the query and term gene lists, their overlap and the statistical significance (P-value) of such enrichment. The user may study the hierarchical relationships between enriched terms in a visual graph view. A typical user input and output of the g:Profiler analysis is shown in Figure 1.
Figure 1.
(A) A typical user input and output scenario of g:Profiler. User inserts a set of genes in the main text window and optionally adjusts query parameters. Results are provided either graphically or in textual format. Genes are presented in columns, and significant functional categories in rows. The analysis of an ordered list shows the length of the most significant query head. GO annotation evidence codes are coloured like a heat map, showing the strength of evidence between a gene and GO term. The legend is provided at the top of the page. It is displayed when the user clicks on the tree icon on the results page. The g:Orth, g:Convert and G:Sorter tools are directly linked to relevant genes from the current query. Additional examples are available in Supplementary Data. (B) Hierarchical relations between the resulting GO categories can be browsed by clicking on corresponding icons.
g:Profiler uses cumulative hypergeometric P-values to identify the most significant terms corresponding to the input set of genes. Unlike most of the common profiling tools mentioned in (10), g:Profiler supports annotations of descendants according to the “True Path Rule” (16). While some tools have adopted GO Slim or level-wise approach to speed up searches and to provide higher-level terms only, the level of abstraction of the output of g:Profiler is entirely determined by the input list and the statistical significance of the matches. Larger queries tend to match more general functional categories and vice versa. Since the scan through every term (GO, pathways, TFBS) may result in many functional annotations, the user must maintain control of what is being reported and what the dependencies between such terms are. g:Profiler provides a GO-structure-preserving visualisation that captures the hierarchical relationships between significantly enriched categories.
Incrementally profiling ordered lists of genes
A significant feature of g:Profiler is the ability to work with ranked or sorted lists of genes. For instance, one may pick any gene of interest and compose such a ranked list by simply sorting other genes by similarity of gene expression. Alternatively, genes may be ranked based on the significant differential expression. The head of such a list should be most informative in determining the functional relationships within these lists. Given an ordered list, g:Profiler incrementally probes all possible sizes of the list head and rapidly determines functional annotations and cut-points in the list where these annotations are most significant. Visualisation shows these cut-points as well as the changes in the P-values as set size increases. The sorted list approach helps to fine-tune the level of abstraction—head of the list may reveal rare specific functions that would otherwise remain insignificant in a large gene set. More precise predictions of putative gene functions can be derived through association to most similar genes. A significant effort has been made to analyse such queries almost as fast as precise flat list queries.
g:SORTER—ranking genes according to expression similarity
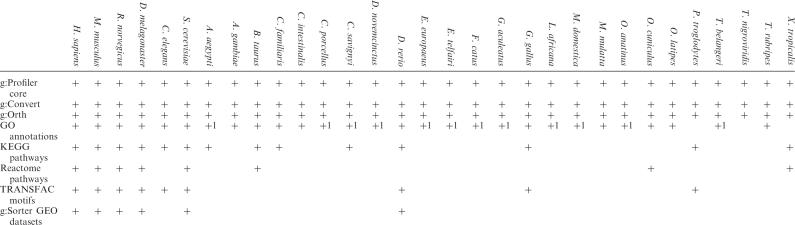
Motivated by user requests for gene-expression-similarity-based analysis, we developed the g:Sorter tool, which allows quick search for similarly or oppositely expressed genes from public data sets downloaded from the GEO database (17). See Table 1 and the web site for a complete overview of the currently supported data.
Table 1.
Overview of the functionality and data sources for different organisms in g:Profiler. Entries with (1) have less than 10 000 related GO associations
Similar genes are often grouped together using clustering, which in many cases depends heavily on initial parameters as the number of expected clusters, for example. With g:Sorter, one can answer questions like ‘What are the 50 most similar probes to gene X?’ or ‘Which probes show reverse regulation in comparison to gene X?’. These direct questions can sometimes be very informative about the gene and its immediate similar neighbours.
The main input of g:Sorter is a single gene ID. The user selects an expression dataset, a mathematical measure of distance like the Pearson correlation or Euclidean distance, and the size of the desired result. The result of g:Sorter analysis is a list of probes most similar (or dissimilar) to the query gene in the selected dataset. Visualisation shows the relative distances between probes. In case a gene is represented by several probes, a search is conducted for each of them. All the derived probe lists are cross-linked to g:Profiler for immediate functional profiling, g:Convert for mapping to other types of database IDs and g:Orth for identifying the orthologs from other species.
g:CONVERT—a guide in the gene namespace maze
Due to historical, organisational, and domain-specific reasons, we need to cope with various databases using different names and identifiers for genes, related proteins, or reporters (probes) of high-throughput platforms. The richness of the naming conventions, identifiers and related mapping procedures form a well-recognised bottleneck of ontological analysis and functional profiling of high-throughput data (10). Conversion from one ID type to another is not a trivial task and often results in loss of information. A gene name in one database may give multiple or no corresponding identifiers in another, and some conversions may be achieved only via a third database. Not only is it hard for users to use different IDs for every separate tool, but also the data underlying these tools comes from various sources with different IDs. GO annotations involve identifiers from organism-specific databases, KEGG database prefers Entrez IDs, transcription factor related data is often available with RefSeq IDs, while input gene lists for profiling usually include microarray probes.
A significant fraction of GO annotation tools currently support only organism-specific naming conventions provided in the GO annotation files or a few mainstream databases. Thus, users may be required to manually match their genes or seek ad hoc solutions through third-party tools. g:Convert supports 31 organisms and allows arbitrary conversions between more than 100 different gene or protein naming schemas, ID types and microarray platforms.
The main input of g:Convert is a list of names, identifiers or accession numbers of genes, proteins and probes from a specific organism. Moreover, the input may be presented as a mix of different ID types. The user only needs to choose the desired output type. g:Convert mappings are based on Ensembl IDs for genes, transcripts and translations via a three-level index. The output of g:Convert is a tabular representation of the input list with corresponding entities in target schema, their gene symbols or names and short descriptions. A simple text-based output is also provided that can be exported to spreadsheet format or integrated with external tools via the HTTP protocol. The same conversions are also used by other modules of the g:Profiler suite. When performing gene list profiling or orthology search, for example, modules automatically query g:Convert and retrieve suitable gene name translations.
g:ORTH—identifying orthologous genes from other species
A lot of insight into biology can be gained only in relation to evolution. For instance, many GO annotations for Homo sapiens are based on predictions on similar genes in model organisms. For an interesting set of differentially expressed mouse genes, it may prove beneficial to retrieve their human and rat orthologs in order to compare expression properties or identify common transcription factor binding sites (12).
Although several bioinformatics groups have created ortholog mappings, there is a lack of easy-to-use tools for end users. The g:Orth module is intended to make these mappings easily available. The orthology data has been downloaded and is regularly updated from the Ensembl database (18).
A user of g:Orth is expected to simply insert a list of genes in one species and select the target organism. Output is a table, where rows display input genes and their corresponding orthologs with names and descriptions, as well as other organisms that have the same ortholog present. A simple text-based output is also provided; this can be exported to spreadsheet format or integrated with external tools via the HTTP protocol.
g:Profiler resources
g:Profiler supports 31 different organisms and offers interface for a list of different databases for functional classification, as well as tasks like namespace conversion, expression analysis, and orthology search (Table 1). g:Profiler resources are kept up-to-date monthly as new Ensembl versions are released. A short description of all sources is given below.
Genomes for 31 organisms, respective namespace mappings, orthology matches, and GO annotations are all retrieved from the Ensembl database (18) via the BioMart (19) interface. We keep our sources up to date and add new organisms as data becomes available. Since Ensembl lacks mappings for a noticeable amount of microarray probes, we fetch additional microarray probeset data from the Gene Expression Omnibus (17).
GO is the primary resource for annotating gene groups to three types of knowledge—cell components (cc), molecular functions (mf) and biological processes (bp) (5). GO is a structured vocabulary in a form of a directed acyclic graph. Hierarchical relations hold within GO; vocabulary terms are related to one or several more general ‘parent’ terms. Any term automatically involves all terms below via all relational paths. Therefore, genes annotated to a specific term in g:Profiler are also added to all associated ‘parents’, and the profiling is performed at all hierarchical levels simultaneously. g:Profiler strips out GO annotations that apply the ‘NOT’ qualifier.
KEGG database provides g:Profiler with functional annotations for metabolic and information processing pathways, cellular processes, human diseases and drug development data (13). KEGG classifications are available for 15 organisms.
Reactome is a mammalian-specific pathway database with thorough annotations of numerous well-studied biological processes, ranging from intermediary metabolism to signal transduction to cell cycle and apoptosis (14). Reactome annotations are available for eight organisms.
Putative transcription factor binding sites from TRANSFAC database (15) are available for nine organisms and retrieved into g:Profiler through a special prediction pipeline. First, TFBS are found by matching TRANSFAC position-specific matrices using the program Match (20) on 1000-bp upstream regions, as provided by the UCSC genome database (21). A cut-off value provided by TRANSFAC is then applied to remove spurious motifs. Remaining matches are split into five hierarchical and inclusive groups based on match score. In most cases, motif matches from the deepest hierarchy are perfect representations of the initial matrix. The hierarchical approach allows TFBS profiling in greater detail and allows the user to distinguish between high- and low-credibility matches.
Data representation and visualisation
With growing amounts of available biological data, the researcher's role in understanding, interpreting and verifying results becomes ever more important. Visual coding of information such as charts, tables, graphs and colour schemes offer great aid in situations where data load is beyond human perception, and a few omitted details may influence research course.
We have put great effort in developing the g:Profiler graphical user interface to combine the variety of profiling information into a concise visual package that best summarises input data (Figure 1). A typical result for ontological analysis is a set of matching functional terms from GO, pathways and TFBS that describe the input set of genes. In g:Profiler, results are depicted in a table, where every row corresponds to a matching functional description.
In addition to numerical information (size of query, significant category and their overlap, the P-value of hitting such term), we present a novel visualisation technique we refer to as gene-to-term mapping. For every gene in input query, the mapping shows a coloured box if there is an association with a term in question. Such a visual solution gives the researcher better intuition about the results that go far beyond the single value of statistical significance.
According to the GOA (6), about 70% of GO annotations to H. sapiens are based on automatically inferred electronic evidence (IEA). Only a fraction of all annotations are verified through strong experimental means. We believe that it is essential for a researcher to acknowledge and account for different types of evidence. The used colour coding helps to cope with the problem of varying quality of evidence behind GO and other functional annotations (10). We have, therefore, provided colour codes to different evidence codes in heatmap style. Weaker, computationally predicted annotations are depicted in blue, while stronger direct evidence from direct assays or interactions are shown in red or orange. The idea of evidence codes is extended to other terms. Pathways that present well-studied information are shown in black, while putative TFBS that tend to be noisier are shown in light colours. Unrecognised gene products or unknown annotations are easily grasped as they are displayed in grey. Colour-coded evidence codes provide a bird's eye view over the whole input gene set and help to assess how well the genes have been previously studied. Most importantly, it shows exactly which genes are annotated to each category, and which genes or categories are related to each other.
Resulting representation is highly interactive, i.e. users can click on the visualisation to browse the data further. In addition to retrieving genes from the categories and their intersections with the query, the user may also view relationships of significant related terms in automatic graph layout mode (Figure 1B). For sorted-list queries, the statistical significance changes are shown relative to the increasing list lengths. The user may start a new query with genes common to a term and initial query, observe genes and associations in a specific term, convert gene names to a different namespace, or search for orthologs in other organisms. Each functional term is provided with an external crosslink to the native data source.
Besides graphical output aimed at human eye, we have also implemented textual output that can be imported into spreadsheets or retrieved automatically via HTTP for processing by other programs. Unlike several popular functional profiling tools referred in (10), g:Profiler is freely available on the web, requires no registration or login procedures, places no demands on operating system or additional software components and is automatically kept updated.
g:SCS threshold—a solution to multiple testing problem for complex data
A crucial factor in functional profiling is the estimation of statistical significance due to multiple testing against many categories if the specific functional category was not selected a priori (10). The explanation to multiple testing in functional classification context is rather intuitive; as we increase the number of terms (such as GO or TFBS), we tend to see more and better-looking matches that may have in fact occurred by chance. In order to keep the experiment-wide threshold at predefined P = 0.05, we need to consider stronger threshold for every individual test of the experiment.
Multiple testing corrections can broadly be split into two groups. Family-wise error rates (FWER) such as Bonferroni or Šidák measure the chance of at least one false-positive match. The test-wide threshold is achieved by decreasing the individual test P-values (i.e. P = 0.05) according to the number of performed tests. Functional profiling provides testing against hundreds to thousands of terms, and such approaches become rather conservative, especially as tests are not independent due to the hierarchical structure of GO. A more liberal group of corrections, false discovery rates (FDR), measure the proportion of false discoveries in a multi-test experiment and gain a test-wide threshold by ranking observed P-values and comparing their relative rank to individual test thresholds (22).
Multiple testing issues are well acknowledged and discussed in functional profiling community (23–25). However, the current situation is far from clear and intuitive. Corrections such as Bonferroni are designed for testing on multiple independent tests, which surely does not apply for heavily overlapping functional classifications from GO. FDR approaches are more promising, since some versions also allow partial dependencies in input data (26). It is not yet known whether the GO hierarchy complies with those schemas. Most profiling tools mentioned in (10) provide either Bonferroni or independent FDR correction, other tools as Onto-Express (8) or FatiGO (23), for example, allow the user to select from multiple schemas, leaving the whole responsibility to the end user, who often prefers to apply the default selection.
In g:Profiler, we introduce g:SCS (Set Counts and Sizes), a novel method specially developed to estimate thresholds in complex and structured functional profiling data such as GO, pathways and TFBS, where statistical significance is determined from set intersections in 2 × 2 contingency tables. We first performed extensive simulations with nearly 10 million randomly sampled simulation queries for different organisms in order to observe the distribution of best P-values for different input query sizes. As a result, an analytical approach was derived that estimates reasonably well the P-value threshold α for which there is 95% chance of having no significant results for a random query. Using binary search, we cover all possible values of α in the range 0–1 with precision 1E − 6. For any such α, we perform the following steps:
For every term, we calculate the probability that a random query would have an enrichment P-value at least α.
We estimate the overall probability of having no significant results for a random query as the product of single probabilities from the previous step.
Next α will be smaller or greater, depending on if the overall probability was smaller or greater than 95%.
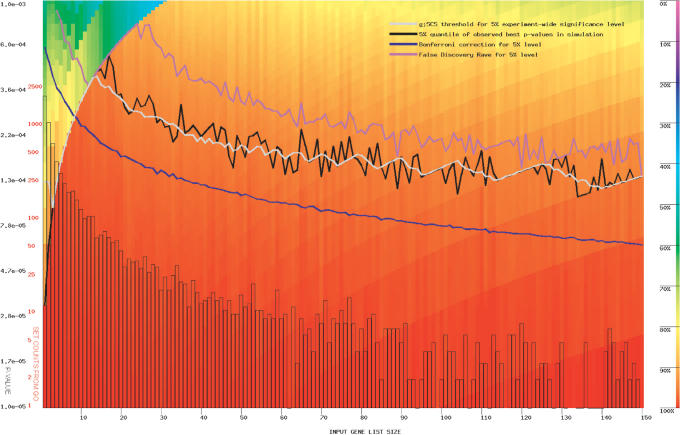
Significance thresholds from the g:SCS method are well verified with simulations; the 5% quantile of best P-values from randomly sampled queries perfectly agrees with values from g:SCS. To compare the g:SCS with empirical randomisation-based significance, we conducted 2000 random queries of every possible size. A single best P-value was recorded for every query and a 5% quantile was determined for each length. The 5% quantile denotes the P-value that is smaller than 95% of observed random P-values. To show the other observed P-values, we used colour coding (Figure 2). On the same background, we overlay the Bonferroni correction, the FDR implied cut-offs and our new g:SCS estimation. Clearly, the g:SCS is able to capture the empirical estimation of the significance of the single test throughout the full scale of the query much better. The jumps in empirical as well as g:SCS thresholds can be visually correlated to the underlying set structure that we have illustrated with the histogram showing the number of gene sets of every different size.
Figure 2.
Comparison of multiple testing corrections for H. sapiens GO annotations for 2000 randomly generated queries for each query size between 1 and 150. X-axis shows the input query size, and Y-axis represents a P-value of the significance threshold of the single set comparison. Lines represent the thresholds for estimated significance cut-off. Frequency histogram of GO annotation set sizes is shown in logarithmic scale. The SCS threshold (white line) follows the 95% quantile of empirically observed simulated values (black line) very closely. In comparison, the more conservative Bonferroni (blue line) or FDR (purple line)-based estimations are shown. Besides the empirical estimation of the 95% significance level, the colour coding on the background shows how frequently at least one such P-value has been achieved from the randomly generated queries.
We consider g:SCS superior to standard multiple testing methods, since it takes into account the actual structure behind functional annotations. The general idea with corrections of 5–15% to global P-value applies to several tested organisms, including H. sapiens (Figure 2), M. musculus, and S. cerevisiae. g:Profiler uses g:SCS thresholds by default. The user interface also allows using the Bonferroni or FDR corrections or any user-given threshold. In case of multiple active schemas, the strongest correction is applied for every single case. If user-given threshold is below statistical significance levels, the respective entries are shown in grey.
CONCLUSION
Since the original development of the GO resource, it has become the de facto standard for studying and comparing functional data of many different organisms. The more we know about the possible function of each and every biological entity, the better we can interpret the complex biological phenomena that we try to dissect with high-throughput studies. In such studies, we need to interpret the relationships between hundreds if not thousands of genes simultaneously. This analysis must be made simple yet powerful to offer the best value for end users. We have made an attempt to simplify the functional analysis process of gene lists and orders, by developing fast algorithms and highly visual representations of the analysis results. The use of the tools has been made simple by supporting many organisms and many more gene-naming conventions in the same user interface. Translating between these ID types and even finding orthologous genes from other species is integrated seamlessly to help the typical end users. With the means to search hundreds of public data sets the exploratory analysis for genes has been made simpler.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work has been supported by the EU FP6 grants ENFIN LSHG-CT-2005-518254, ATD LSHG-CT-2003-503329 and FunGenES LSHG-CT-2003-503494; University of Tartu base funding for establishing the research group and Estonian Science Foundation grants 5722 and 5724. The authors would also like to thank the anonymous referees for useful comments that helped to improve the final version. Funding to pay the Open Access publication charges for this article was provided by University of Tartu.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Eads CA, Daneberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Daneberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. PNAS. 2001;98(8):4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83(3):349–60. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32(1):D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Shahrour F, Minguez P, Vaquerizas J, Conde L, Dopazo J. Babelomics: a suite of web-tools for functional annotation and analysis of group of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz S, Tainsky MA. Onto-Tools, The toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31(13):3775–81. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 10.Khatri P, Drăghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21(18):3587–95. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-lalpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 12.Lehner B, Fraser AG. A first-draft human protein-interaction map. Genome Biol. 2004;5:R63. doi: 10.1186/gb-2004-5-9-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi-Tope G, Gillespie M, Västrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–32. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Creating the Gene Ontology Resource: Design and Implementation. Genome Res. 2001;11(8):1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasprzyk A, Keefe D, Smedley D, London D, Spooner W, Melsopp C, Hammond M, Rocca-Serra P, Cox T, Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kel AE, Gößling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCHTM: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat. Soc. 1995;57:289–300. [Google Scholar]
- 23.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 24.Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19(18):2502–4. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 25.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.