Abstract
OSCAR is a web platform for cluster and cross-species analysis of microarray data. It provides a comprehensive but friendly environment to both users and algorithm developers. For users, OSCAR provides cluster tools for both single and multiple species data, together with interactive analysis features. For single species data, OSCAR currently provides Hierarchical Clustering, K-means, partition around medoids (PAM), Self-Organizing Map (SOM), Tight Clustering and a novel algorithm called ‘Consensus Tight-clustering’. The new Consensus Tight-clustering algorithm delivers robust gene clusters and its result is more resistant to false positives than other state-of-the-art algorithms. For cross-species data analysis, OSCAR provides two novel computational tools: ‘coherentCluster’, ‘coherentSubset’ and a novel visualization tool: ‘comparative heatmap’. Applying the coherentCluster algorithm to human and fly aging data, we identified several coherent clusters of genes, which share co-regulation patterns that are highly correlated with the aging process in both of the two species. One coherent cluster suggests insulin receptor (INSR) may regulate Pax6 in both species and across different tissues. Further analysis with human brain expression and pathological data suggests an INSR->Pax6->quinone oxidoreductase (NQO1)->detoxification neuro-protective pathway might be present in aging or diseased brain. For algorithm developers, OSCAR is a plug-and-play platform. With little effort, developers can plug their own algorithms into the OSCAR server without revealing the source codes, which will equip their command line executables with user-friendly interface and interactive analysis capability. In summary, OSCAR initiates an open platform for development and application of clustering and cross-species analysis programs. OSCAR stands for an open system for cluster analysis of microarray data. It is available at: http://biocomp.bioen.uiuc.edu/oscar
INTRODUCTION
Microarray technology enabled simultaneous quantification of the expression of thousands of genes (1,2). Functional relevance among genes, such as sharing a common transcriptional regulator protein or responding to a common regulatory signal, may induce co-expression: a group of genes sharing a similar expression pattern over time or across different conditions (3). Although it does not necessarily imply a causal relationship among transcript levels, co-expression has been shown in a number of studies to be correlated with functional relationships (4–7). The identification of co-expression gene groups can lead to identification of common regulatory motifs (8,9), inference of signaling pathways (10) and genetic networks (3).
Cluster analysis, an unsupervised learning method for identification of co-expression groups, is commonly used as an initial investigative tool of microarray data before specific pathways or genetic mechanisms are scrutinized (3–6,10). When genes are subject to cluster analysis, a total of ‘n’ genes are assigned into ‘K’ clusters of similar expression patterns given a dissimilarity measure between any two genes. The appropriate choice of the dissimilarity measure, i.e. a distance metric for cluster analysis, is arguably as important as the choice of the clustering algorithm itself (11), although the importance of the latter choice is more obvious to users. A relatively comprehensive evaluation of various gene-clustering methods has been carried out recently (12), and interested readers are referred to this article for detailed comparison. A selection of clustering methods that are commonly used in microarray analysis is categorized subsequently. Non-parametric algorithms hierarchical clustering (4), K-means (13), partitioning around medoids (PAM; a.k.a. K-medoids) (14), self-organizing maps (SOM) (15,16), fuzzy K-means (17), consensus clustering (18) and tight clustering (19) and robust multi-scale clustering (20) are among the most popular ones. Among these algorithms, fuzzy K-means differs from others in that it allows a gene to belong to multiple clusters. Tight clustering differs from others in that it does not require every gene to fall into a cluster, but allows scatter genes: genes should not be assigned into any clusters. Tight clustering and robust multi-scale clustering share the same idea of using co-occurrence matrix from multiple runs. Model-based algorithms (21–27) usually assume that data come from a Gaussian mixture model, but can also allow a component of homogeneous Poisson process for scattered genes (28). To avoid subjective dictation of the number of clusters, Chinese Restaurant Process, a mixture of countably infinite number of simple distributions is recently applied to analyze microarray data (29) [Chinese Restaurant Process is elegantly reviewed by Michael Jordan (30)]. Further developments on clustering related analysis include bi-clustering, which identifies co-expression in subsets of samples (31–33), second-order correlation, which groups gene-duplets instead of single genes (34), and co-expression dynamics, which simultaneously models gene expression and time-dependent cellular state (35).
The aforementioned is only a partial list of the clustering algorithms available for analyzing microarray data. Although quite a few software tools each implemented one to several algorithms (4,36–41) (Supplementary Table S1), not a single software tool has incorporated the majority of available algorithms. Especially the recently developed algorithms are usually not incorporated into any software with a friendly user interface. The problem for cluster analysis is that there is no best algorithm. An algorithm can work better for some data sets but not as good on other data sets. The visualization in one software tool is usually different from that of another tool, which makes it difficult to compare the results unless to scrutinize the text output for each cluster. A system that allows users to run multiple algorithms side by side and deliver comparable visualization results will be tremendously valuable to microarray data analysis. The OSCAR system allows this to happen.
METHODS
The OSCAR system
The OSCAR server and web application is an open system for cluster analysis of microarray data, with an automated procedure to incorporate and manage all clustering algorithms. It provides a comprehensive and friendly environment to both users and algorithm developers. A database system is developed to manage all the algorithms, including their documentation, their parameters, each parameter's description, type, bounds and default value. When a user accesses the OSCAR website, the server will automatically list all the algorithms currently available, together with a URL to the documentation for each of the algorithms listed. When a user chooses a particular algorithm, all information about the parameters and input files of the algorithm is retrieved from the algorithm database and automatically displayed to the user. Users can use the interactive web forms to adjust the parameters, upload input data and execute the computation on the server. Algorithm ‘developers’ can use the interactive web forms to incorporate their own algorithms to OSCAR without revealing their source codes. The submitted algorithm will be managed by OSCAR's database, sharing the same output format and be accessible to all users (Supplementary Figure S1).
OSCAR for users
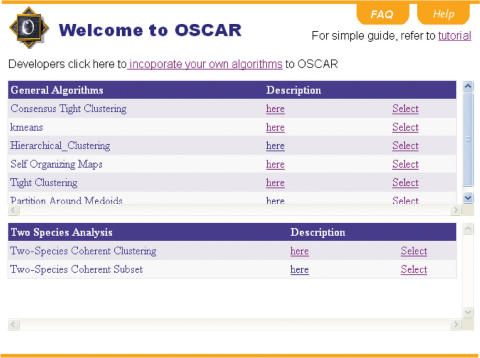
OSCAR provides an intuitive web interface to users. When a user accesses the OSCAR main page, all currently available algorithms and hyperlinks to their documentations will be retrieved from the algorithm database and displayed (Figure 1). The user can select any algorithm listed. Upon selection, the specifications and default values for all the parameters required by the user-selected algorithm will be retrieved from the database and displayed to the user (Supplementary Figure S2). The user can modify the default parameters, upload input data and execute the computation. Sample inputs files are provided for user's convenience. Some users may want to quickly try out each algorithm and get a sense of what each one is doing. This can be achieved by clicking the ‘Submit using sample files’ button. Output is provided to users in two formats: text (Supplementary Figure S3) and interactive heatmap (Figure 2). The user can save both outputs to a local computer by clicking the disk icon in the upper right corner of the web page. Users can alter the color schemes used by the heatmap by clicking the ‘Change Color’ button. Two schemes are provided: red-green and blue-yellow. Hovering mouse cursor over sample names or any spot within the heatmap will invoke a small pop-up window next to the cursor, containing either information about the sample or the gene expression value used to draw the color in the cursor covered area.
Figure 1.
Screenshot of the main web page for users.
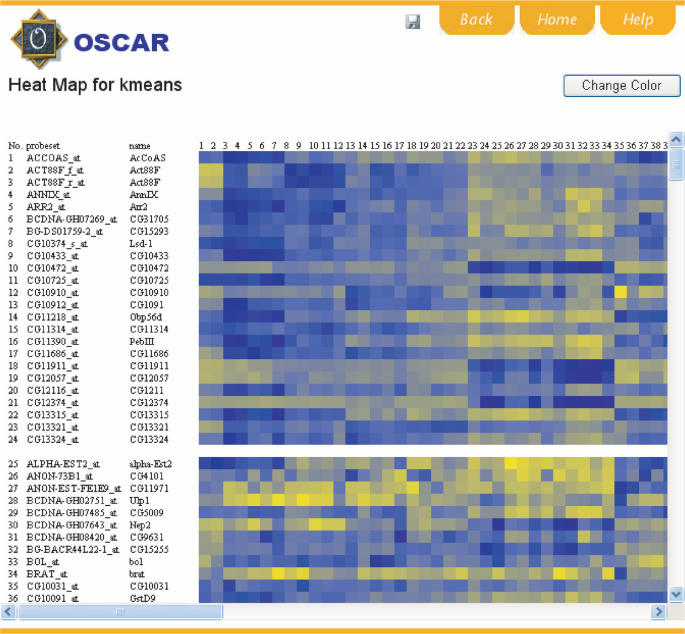
Figure 2.
Screenshot of a heatmap output. The heatmap is interactive. Hovering mouse cursor over each spot on the heatmap will dynamically retrieve the gene expression values. Hovering cursor over sample numbers will retrieve full sample names. Blue represents lower expression and yellow represents higher expression. The color scheme can be changed by clicking the ‘Change Color’ button. The figure can be saved by clicking the disk button.
The algorithms currently available to OSCAR users are: (i) hierarchical clustering, (ii) K-means, (iii) partition around medoids (PAM), (iv) self-organizing map (SOM), (v) tight clustering, (vi) Consensus Tight-clustering (new), (vii) two-species coherent clustering (new). Users can choose any of the following distance metrics to be used in hierarchical clustering, K-means and PAM: (a) Pearson correlation, (b) absolute value of the Pearson correlation, (c) uncentered Pearson correlation, (d) absolute uncentered Pearson correlation, (e) Spearman's rank correlation, (f) Kendall's τ, (g) Euclidean distance and (h) city–block distance. Three linkage definitions are allowed in hierarchical clustering: (1) single linkage, (2) complete linkage and (3) pairwise average. For the purpose of comparison, hierarchical clustering will give usual co-expression groups as outputs instead of hierarchical trees. This is achieved by trimming the hierarchical tree by the allowed maximum number of clusters provided by the user.
Three new algorithms are delivered to users through OSCAR. One general clustering tool and two two-species analysis tools (will be discussed later). ‘Consensus Tight-clustering’ is a new algorithm that blends the advantages of two very recently published non-parametric clustering algorithms: tight clustering (19) and robust multi-scale clustering (20). It is interesting to notice that these two recent algorithms employ very similar ideas, except that tight clustering is stronger in that it used a re-sampling strategy and robust multi-scale clustering is stronger in that it utilizes the co-occurrence matrix for all individual runs rather than only the runs under consecutive Ks in K-means. The new Consensus Tight clustering algorithm basically adds the re-sampling step to each iteration of the robust multi-scale clustering (Supplementary Figure S4). Compared with multi-scale clustering, Consensus Tight-clustering can identify more robust gene clusters, i.e. is more resistant to inclusion of false positives in a cluster. This is because Consensus Tight-clustering re-samples the data in each iteration, and summary from re-sampled data is more robust than summary from the original data (19). Compared with tight clustering, Consensus Tight-clustering increases both sensitivity and specificity for the following reasons. Due to programming difficulty and consideration of computational efficiency, the tight clustering program made two simplified approximations. First, tight clustering only stores seven largest clusters for each K in the K-means clustering, and second, tight clustering only checks re-occurrence of the seven stored clusters in the results from K-means computation of an immediately larger K. Ideally the program should store as many clusters as possible in each K and cross-check the re-occurrence of these stored clusters across a number of different Ks. Consensus Tight-clustering implements these ideal treatments and therefore improves from tight clustering. In our tests, we also found Consensus Tight-clustering can handle larger input data than tight clustering and is much less likely to break down during the computation. A potential weakness of Consensus Tight-clustering is that it may consume more computational time. In our tests of four real data sets, Consensus Tight-clustering is slightly slower than tight clustering in two data sets, but much faster than tight clustering in the other two data sets (Supplementary Table S2). Although more tests are needed to make the final conclusion, Consensus Tight-clustering seems to improve computational efficiency over tight clustering on an overall scale.
We tested four data sets on the OSCAR server. These data sets are related to human colon cancer (42), mouse pre-implantation time course (43), fruit fly aging with and without calorie restriction (44) and worm natural aging (45). The run time for generating text and heatmap of each algorithm on each data set is recorded (Supplementary Table S2). We acknowledge that the run time for each algorithm is sensitive to the actual parameters used, and Supplementary Table S2 only reflects one set of parameter values out of many possible sets.
Tools for cross-species analysis
Recently a number of interspecies comparisons of gene expression levels have been carried out in various phylogenetic branches, including human and monkeys (46,47), rodents (48), human and mouse (49), Xenopus (50), Drosophila (51) and plants (52). One of the central questions that inspired these studies is how natural selection acts on regulation of gene expression. Most of these studies aim for answering evolutionary questions pertaining to gene expression changes, for example, whether the expression divergence of most genes can be explained by a neutral theory. To the authors’ knowledge, there are no published methods on cluster analysis of multiple species data. OSCAR provides two novel computational programs and a novel visualization tool, comparative heatmap, to facilitate cross-species analysis.
The two cross-species clustering programs are: coherence clustering (coherentCluster) and coherent subset (coherentSubset). Both tools identify clusters in which homologous genes show co-expression in both species. The biological motivation is: if a group of genes show conserved expression patterns in the data set from two different species, then it is very likely that the group is under evolutionary constraint and thus a functionally related group. This will allow one to distinguish between gene clusters with biological significance and the clusters that are consequences of experimental or computational artifacts.
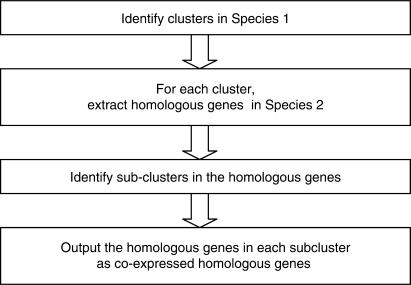
CoherentCluster takes three input data files: one microarray data file for each species and a homologous gene-mapping file. The mapping file should contain two columns, with homologous gene IDs for the two species occupying the same row of the two columns. In the computation, coherentCluster first performs Consensus Tight-clustering in one species, and then for each cluster, coherentCluster extracts the homologous genes in the other species (Figure 3). If the cluster result in the first species were free of false positives, and if all the homologous mapping were correct and the regulation of all the genes were evolutionarily conserved, the homologous genes of a cluster should be clustered in the second species as well. In reality, all of the three assumptions above can be violated, and homologous genes of a cluster can usually break down into a few sub-clusters together with scatter genes in the other species. Each ‘homologous sub-cluster’ represents a group of genes being co-expressed in both of the two species. We term the homologous gene group with co-expression in both of the two species as ‘coherent clusters’. It should be noticed that a coherent cluster does not imply the genes within should have the same expression patterns in two species. As long as the genes form a cluster in both species, they form a coherent cluster (Figure 4). The CoherentCluster program gives coherent clusters as output. Coherent clusters can be viewed in OSCAR by ‘comparative heatmap’ (Figure 4). Finally, on the technical side, there could be multiple ways to identify sub-clusters in the second species. CoherentCluster first samples a large number of gene pairs from all the genes in Species 2, and it derives an empirical distribution of the pairwise distances. The user decides a percentage of the pairwise distance within which two genes should be regarded as co-expressed. For example, the user may designate the fifth percentile of the pairwise distances as the threshold of co-expression. With this threshold, coherentCluster identifies the co-expressed sub-cluster whose homologous genes were also co-expressed (Supplementary Figure S5A for pseudo code).
Figure 3.
Flowchart of the coherentClustering algorithm.
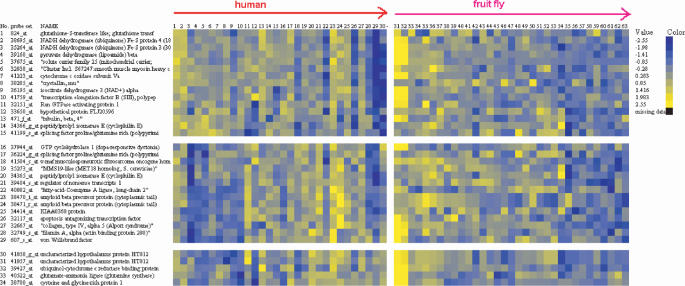
Figure 4.
The ‘comparative heatmap’ showing the result from an analysis of aging data in two species. The displayed probe sets and gene names are retrieved from the second input species. The expression patterns of the homologous genes in the two species are displayed side by side, separated by a vertical bar. Each cluster represents a subset of genes that show coherent expression in both of the two species. The figure is interactive and sample information will appear in a pop-up window if the user hovers the cursor on the sample IDs. Only 3 of the 46 coherent clusters are displayed. See Supplementary Figure S6 for all the 46 coherent clusters.
To identify coherent clusters, users do not have to start from Consensus Tight-clustering in the first species. Instead the user may want to start from any other clustering result or even from other suggestive evidence, such as sharing of similar sequence motifs. The coherentSubset program takes three input files as well, (i) a file of cluster results from one species clustering analysis (the output from any clustering algorithm), (ii) gene expression data from another species and (iii) a homologous gene ID mapping table. CoherentSubset identifies co-expressed subgroups of each input cluster in the other species. CoherentSubset performs the same computation as coherentCluster except that it uses the user defined cluster in the first species to substitute the result from Consensus Tight-clustering (Figure S5B). It splits each user input cluster into two-species ‘coherent subsets’. Each coherent subset contains genes showing co-expression in the second species from the second input file.
CoherentCluster and CoherentSubset use a unified output format — ‘comparative heatmap’ (Figure 4). Comparative heatmap displays the expression of the homologous genes in the same row of a heatmap, using a vertical bar separating the two species. The color on the heatmap is normalized within each species, and therefore the contrast of the color for each species represents the change of gene expression within that species. The spacing used to separate the clusters can be small or large. Small spacing separates the clusters that show coherent pattern in one of the two species. Large spacing separates the clusters with different patterns in both species. The result should be interpreted as follows: if the user only had one species data, she/he would obtain clusters separated by large spacing. Each cluster contains co-expressed genes in this species, but their homologous genes in the other species may not all be co-expressed. With the second species data, each cluster further breaks down into smaller clusters, separated by small spacing. Each small cluster represents a group of genes show co-expression patterns in both of the two species (Figure 4).
To illustrate the use of the two species analysis algorithms, we give a data example using the coherentCluster algorithm. There have been a few competing hypotheses regarding the aging process. First, do different tissues within an animal share the same aging program? Or do they each possess their own aging programs? Second, do phylogenetically highly diverged species share a core set of genes whose expression is correlated with their aging processes? Analyzing human and chimpanzee gene expression data in different parts of the brain, Fraser et al. (53) found aging is heterogeneous among different regions of the human brain, and chimpanzee cortex ages differently from human cortex. These findings suggest aging is reflected by both tissue-specific and species-specific expression patterns. Rodwell et al. (62) found human kidney possesses organ-specific mechanisms and pathways in its aging process, but two different tissues in kidney do have similar expression profiles. However, McCarroll et al. (54) reported similarly correlated regulation during aging in microarray data sets from different tissues and suggested the existence of a core set of age-related genes across different species, in particular, gene expression in heads of young and adult male files are highly correlated with that of aging worms.
To further investigate the hypotheses regarding shared versus specific gene expression patterns across tissues and across species, we applied the Consensus Tight-clustering and coherentSubset algorithms to gene expression data in the aging processes of human brain (55) and fly whole-body (44). Fly and human are estimated to have diverged for about 600 million years, which was after the divergence of worm and fly (56–59). The human data consist of samples from frontal pole of 30 individuals, ranging from 26 to 106 years of age. The fruit fly data consist of 34 arrays measuring flies from 3 to 47 days of age. Fly samples without calorie restriction were used because they represent the natural aging process as the human samples (calorie-restricted flies were excluded). Both of the two studies used Affymetrix GeneChip microarrays. The data were normalized by the original authors, and we made log2 transformation to all the normalized values. Homologous gene ID map was downloaded from Affymetrix website. Users are also referred to the Ensembl database (http://www.ensembl.org) and its accompanying Biomart software (60), with which homologous gene mapping can be easily obtained. Genes that do not have homologous genes in the other species are removed from the analysis. 2934 fly probe sets and their 3445 human homologous probe sets were retained. (Neither of our two-species algorithms requires this step. We did so to trim down the total number of genes in the analysis.) We first submitted the fly data to Consensus Tight-clustering with default parameters, and identified nine clusters. Next, we submitted the nine fly clusters, the homologous ID map, and the human expression data to the coherentCluster program. With default parameters, the nine fly clusters broke down to 46 coherent clusters (Supplementary Table S3 and Figure S6, Figure 4). We could have achieved the same result by directly submitting the ID map, fly and human expression data to the coherentCluster program, however, with a two-step analyses we also obtained the intermediate results. It is interesting that the three coherent clusters in Figure 4 came from the same fly cluster, with high expression at an early age and the expression decreases over time. However, the homologous genes in human break into three subsets (coherent clusters). The first coherent cluster has a consistent decreasing pattern in human as well. The second coherent cluster has a Λ-shape, first increasing with age and then decreasing in very old individuals. The third coherent cluster has a completely reversed expression pattern in human—increasing over time. These coherent clusters represent groups of genes possibly co-regulated in both species, but with different mechanism of transcriptional control.
INSR->Pax6->quinone oxidoreductase (NQO1) pathway
Insulin receptor (INSR) is contained in one of the coherent clusters, which had increasing expression pattern in both human brain and whole-body flies. This coherent cluster only contains four other genes: Pax6, RhoA, VPS13B and YTHDF3 (Supplementary Table S3, Figure 5). It is well known that mutations in the gene encoding insulin-like growth factor receptor alter lifespan in worms, flies and mice, indicating that an endocrine signaling pathway has a conserved role in aging (61). However, the mechanism with which insulin affects lifespan remains largely unknown. Somewhat counter-intuitively, the expression levels of insulin receptor in human brain and kidney are reported to be positively and negatively correlated with age, respectively (55,62). Because the cross-species analysis assumes divergent species data may help to filter out noisy data, it is natural for us to follow-up the other genes in the INSR containing coherent cluster to explore potential upstream and downstream factors and co-factors relating to the insulin pathway during the aging process.
Figure 5.
The insulin receptor containing coherent cluster.
Pax6 encodes a transcriptional regulator involved in developments of sensory organs, central nerve system and pancreas. Insulin was known to be primarily produced by β-cells of the islets of Langerhans, the functional units of the endocrine pancreas. β-cells form the core of the islet, whereas α-cells are arranged at the periphery of the islet and secrete glucagon. The Pax6 gene is expressed during the early stages of pancreatic development and in mature endocrine cells and it is essential for the differentiation of α-cells (63).
The fact that Pax6 is one of the only four genes consistently co-expressed with insulin receptor provokes a hypothesis—Pax6 is regulated by insulin signaling in developing and adult tissues: in developing pancreas, Pax6 in response to insulin produced by neighboring β-cells, controls the development of surrounding α-cells; in adult brain, Pax6 responds to insulin receptor and carries out its effect by transcriptionally regulating other genes. In other words, we hypothesize that Pax6 is a downstream regulator of an insulin receptor mediated genomic pathway. Although to the authors’ knowledge Pax6 has not been shown to be responsive to insulin pathway, mutation of Pax6 can lead to glucose intolerance and early onset of diabetes mellitus (64,65). Pax6 is also required to achieve insulin-dependent inhibition of the transcription of the glucagon gene in a pancreatic islet cell line (66). The co-expression of Pax6 and insulin receptor in both human brain and whole fly may suggest a phylogenetically conserved pathway, and it could be utilized by multiple tissues (recall the hypotheses that inspired our analysis.)
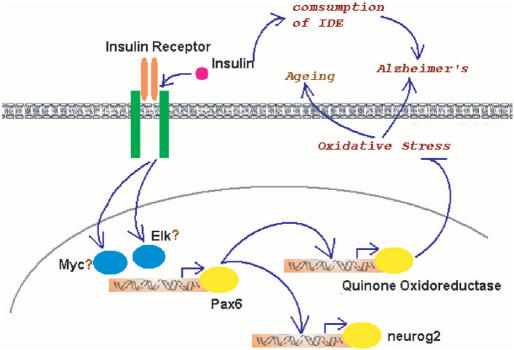
We further investigated the potential effects of the INSR->Pax6 pathway in human brain. Neurogenin-2 and NQO1 are two genes known to be transcriptionally regulated by Pax6 in spinal cord and in the eye, respectively (67,68). This led us to investigate whether NQO1 is also regulated by Pax6 in the brain (Neurogenin-2 is not represented on the microarray). Not surprisingly, the expression of NQO1 is strongly correlated with that of Pax6 (P-value = 0.007, Supplementary Figure S7). The computationally suggested link of Pax6 bridging insulin signaling to NQO1 in human brain is interesting in 3-folds. First, Alzheimer's disease (AD) is associated with major impairments in insulin signaling in the brain and is reversible by early treatment of insulin sensitizer (69). Second, oxidative stress and stress-activated signaling pathways were found to be strongly associated with insulin resistance (70). Third, NQO1 detoxifies quinones. Quinones are highly redox-active molecules which often lead to formation of reactive oxygen species (ROS), the primary sources of oxidative stresses (71,72). These pieces of evidence, together with the fact that Pax6 and insulin receptor turn up in a coherent cluster, assemble a picture of a neuro-protective pathway, i.e. insulin receptor -> Pax6 -> NQO1 -> detoxification and reduction of oxidative stress -> stop of AD (Figure 6). In line with the argument that NQO1 stands in a self-protective pathway, NQO1 expression was found to regionally co-localize with the pathology of AD (73).
Figure 6.
A hypothetical model for the dual roles of insulin signaling in human brain.
The insulin receptor-mediated genomic pathway may also be a self-defense mechanism against the natural aging process in the brain. Lu et al. (55) showed that DNA damage is remarkably increased in the promoters of genes with reduced expression in the aged brain, and the same promoters are selectively damaged by oxidative stress in cultured human neurons. Oxidative as well as other stresses are known to be mediated at least in part by the production of ROS (61). The INSR->Pax6->NQO1 pathway may therefore defend against aging by inhibiting ROS formation (Figure 6).
In contrast to this new pathway, INSR homologue in worm (daf-2) was known to repress another ROS detoxification pathway, through inhibition of daf-16, a homologue of the HNF-3/forkhead transcription factor, which activates ROS detoxification enzymes (74). To summarize, INSR-mediated genomic pathways may have two blades, one inhibiting and the other activating detoxification enzymes, which exert opposite effects on aging. It is worth pointing out that insulin may also play dual roles in AD. The neuro-protective genomic pathway coexists with a harmful non-genomic pathway. Insulin boosts β-amyloid by monopolizing the attention of insulin-degrading enzyme (IDE) that degrades and clears them both. β-Amyloid is a peptide thought to be the active ingredient in most AD pathology. The more insulin, the less available IDE in the brain, and the higher risks for accumulation of β-amyloid, which leads to AD (75–77) (Figure 6).
RhoA (ras homolog gene family, member A) is another gene co-clustered with insulin receptor. RhoA is a small G-protein in the Rho family that binds to Rho-kinase. Rho-kinase is shown to associate with insulin receptor and inhibits insulin signaling in multiple cell types (78–80), and thereby Rho/Rho-kinase is suggested to be involved in the development of insulin resistance in diabetes (81). The co-expression of RhoA and insulin receptor may suggest an inhibiting mechanism to insulin signaling in multiple tissues (recall we used human brain and fly whole-body data, and also recall the hypotheses that inspired our analysis). Interestingly Rho/Rho-kinase may also be a double-edged sword for inhibition of signaling in human brain. Besides association with insulin receptor, Rho/Rho-kinase activation can suppress insulin gene transcription (82), and brain is indeed an insulin-producing organ (69).
The other two genes in the insulin receptor cluster, VPS13B and YTHDF3, have only recently been cloned and there is so far little functional information available to them. The interesting biological links for the other three genes (INSR, Pax6, RhoA) in this coherent cluster provokes the question of what these two new genes do? The rest of the coherent clusters, although not analyzed in this article, also deserve further investigation.
OSCAR for ‘developers’
OSCAR requires little programming effort for ‘developers’ to add their own algorithms to the system. It takes two steps to submit an algorithm. Step 1: clicking ‘Developers click here to incorporate your own algorithms’, developers will be linked to a web form for submission. Developers should provide the following information on the web form: algorithm name, the location of the executable program (.exe) of the algorithm, a ‘readme’ file (.html or. txt), the number of parameters, the number of input files and an email for contact. After filling the form, the developer can click the ‘Add’ button to proceed to step 2. A dynamically generated web form will appear, requesting more information for each parameter and each input file (Supplementary Figure S8). In this step, the developer should specify the name and provide a short description for each parameter and each input file. The default value, upper and lower bounds for each parameter should also be specified. For each input file, the developer should provide a next file name, a short description and a sample input file. Clicking the ‘Add’ button will invoke a checking process and submit the algorithm. A red star will appear next to any required field that the developer forgets to fill in, and a red exclamation mark will appear next to any field filled with an obviously wrong value, for example, a string is provided as the default value for a parameter of ‘double’ type. If the checking process is successful the algorithm will be added to the OSCAR server and a thank you message will appear.
At this stage this new algorithm is already managed by the algorithm database in OSCAR, however, the database will not release the added executable to user interface until it passes a more rigorous test. This testing step is necessary for two reasons. First, the developer may have submitted a program with errors, for example a non-functional program or that the developer mistakenly specified one less parameter than the program actually needs. Second, for safety to both the server and the users, the submitted programs should be subject to virus scan before users can execute them. The administrators of OSCAR are committed to perform these tests within one week of a submission. The administrators will modify the ‘enabled’ field for the new algorithm from zero to one in the database to release the algorithm. Each time a user accesses the server, only and all of the ‘enabled’ algorithms will be exposed on the web page.
The checking process before enabling an algorithm is necessary and cannot be replaced by automated procedures. OSCAR administrators will verify the developer's identity and communicate with the developer by email. We retain the right to not enable any programs submitted by developers lacking a traceable identity in the research community.
Comparison to other systems
Supplementary Table S1 summarizes other software tools that perform cluster analysis. OSCAR has the following advantages over these systems. First, OSCAR is web based. It does not require installation and has a much shorter learning period for biologists (several minutes) as compared with many other systems. Second, for algorithm developers, it requires much less efforts to contribute a clustering algorithm to OSCAR (basically providing the executable and documentation), as compared with for example learning the R language and writing an R package, which may take one to eight weeks for experienced programmers. Not to say that the majority of other systems do not allow third-party algorithms. Third, OSCAR supports more algorithms than most of other systems, with only exceptions to the systems that require programming capability in the analysis, such as R. OSCAR's target users are biologists and medical scientists, who are unlikely to have programming capabilities. Finally, OSCAR is pioneered in computational tools and features for cross-species analysis. These cross-species clustering tools are most useful when a researcher needs to obtain very reliable gene clusters for further analysis, for example, for identification of transcription factor binding sites. The criterion of conservation of expression patterns will filter out quite a number of false gene clusters.
CONCLUSION
OSCAR is an open system that bridges the gap between the many described clustering algorithms and the few implemented into software tools with friendly user interfaces. The unsupervised nature of cluster analysis demands researchers to try out many clustering algorithms and choose the one that works best for each particular data set. With a unified algorithm management system and unified outputs, OSCAR enables easy incorporation of all clustering algorithms, and allows users to compare them side by side. OSCAR has been tested under Mozilla Firefox, Safari and Internet Explorer running on MacOS, Linux and Windows systems.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Drs. Gene Robinson and H. Rex Gaskins for wonderful suggestions. We thank Drs. M. Hoon, S. Imoto, J. Nolan, S. Miyano, G. Tseng, C. Kim and S. Kim for sharing their source codes. S. Z. is supported by Illinois Institute of Regenerative Medicine and Stowers Institute of Medical Research. Funding to pay the Open Access publication charges for this article was provided by Department of Bioengineering, University of Illinois at Urbana Champaign.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schena M, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 1999;21(Suppl. 1):33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 3.Lee HK, et al. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14:1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramaswamy S, et al. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes DR, et al. Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- 7.Yuen T, et al. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 2002;30:e48. doi: 10.1093/nar/30.10.e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence CE, et al. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 9.Conlon EM, et al. Integrating regulatory motif discovery and genome-wide expression analysis. Proc. Natl Acad. Sci. USA. 2003;100:3339–3344. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JM, Sonnhammer EL. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–882. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim RS, Ji H, Wong WH. An improved distance measure between the expression profiles linking co-expression and co-regulation in mouse. BMC Bioinformatics. 2006;7:44. doi: 10.1186/1471-2105-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thalamuthu A, et al. Evaluation and comparison of gene clustering methods in microarray analysis. Bioinformatics. 2006;22:2405–2412. doi: 10.1093/bioinformatics/btl406. [DOI] [PubMed] [Google Scholar]
- 13.Hartigan JA, Wong MA. A K-means clustering algorithm. Appl. Stat. 1979;28:126–130. [Google Scholar]
- 14.Kaufman L, Rousseeuw P. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley; 1990. [Google Scholar]
- 15.Holdaway RM, White MW. Computational neural networks: enhancing supervised learning algorithms via self-organization. Int. J. Biomed. Comput. 1990;25:151–167. doi: 10.1016/0020-7101(90)90006-g. [DOI] [PubMed] [Google Scholar]
- 16.Erwin E, Obermayer K, K. Schulten K. Self-organizing maps: ordering, convergence properties and energy functions. Biol. Cybern. 1992;67:47–55. doi: 10.1007/BF00201801. [DOI] [PubMed] [Google Scholar]
- 17.Gasch AP, Eisen MB. Exploring the conditional coregulation of yeast gene expression through fuzzy K-means clustering. Genome Biol. 2002;3:RESEARCH0059. doi: 10.1186/gb-2002-3-11-research0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swift S, et al. Consensus clustering and functional interpretation of gene-expression data. Genome Biol. 2004;5:R94. doi: 10.1186/gb-2004-5-11-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng GC, Wong WH. Tight clustering: a resampling-based approach for identifying stable and tight patterns in data. Biometrics. 61:10–16. doi: 10.1111/j.0006-341X.2005.031032.x. [DOI] [PubMed] [Google Scholar]
- 20.Grotkjaer T, et al. Robust multi-scale clustering of large DNA microarray datasets with the consensus algorithm. Bioinformatics. 2006;22:58–67. doi: 10.1093/bioinformatics/bti746. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh D, Chinnaiyan AM. Mixture modelling of gene expression data from microarray experiments. Bioinformatics. 2002;18:275–286. doi: 10.1093/bioinformatics/18.2.275. [DOI] [PubMed] [Google Scholar]
- 22.McLachlan GJ, Bean RW, Peel D. A mixture model-based approach to the clustering of microarray expression data. Bioinformatics. 2002;18:413–422. doi: 10.1093/bioinformatics/18.3.413. [DOI] [PubMed] [Google Scholar]
- 23.Medvedovic M, Sivaganesan S. Bayesian infinite mixture model based clustering of gene expression profiles. Bioinformatics. 2002;18:1194–1206. doi: 10.1093/bioinformatics/18.9.1194. [DOI] [PubMed] [Google Scholar]
- 24.Medvedovic M, Yeung KY, Bumgarner RE. Bayesian mixture model based clustering of replicated microarray data. Bioinformatics. 2004;20:1222–1232. doi: 10.1093/bioinformatics/bth068. [DOI] [PubMed] [Google Scholar]
- 25.Ng SK, et al. A mixture model with random-effects components for clustering correlated gene-expression profiles. Bioinformatics. 2006;22:1745–1752. doi: 10.1093/bioinformatics/btl165. [DOI] [PubMed] [Google Scholar]
- 26.Reverter A, et al. A mixture model-based cluster analysis of DNA microarray gene expression data on Brahman and Brahman composite steers fed high-, medium-, and low-quality diets. J. Anim. Sci. 2003;81:1900–1910. doi: 10.2527/2003.8181900x. [DOI] [PubMed] [Google Scholar]
- 27.Yeung KY, et al. Model-based clustering and data transformations for gene expression data. Bioinformatics. 2001;17:977–987. doi: 10.1093/bioinformatics/17.10.977. [DOI] [PubMed] [Google Scholar]
- 28.Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. JASA. 2002;97:611–631. [Google Scholar]
- 29.Qin ZS. Clustering microarray gene expression data using weighted Chinese restaurant process. Bioinformatics. 2006;22:1988–1997. doi: 10.1093/bioinformatics/btl284. [DOI] [PubMed] [Google Scholar]
- 30.Jordan M. Nonparametric Bayesian Methods: Dirichlet Processes, Chinese Restaurant Processes and All That. NIPS. 2005 [Google Scholar]
- 31.Cheng Y, Church GM. Biclustering of expression data. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:93–103. [PubMed] [Google Scholar]
- 32.Qu H, et al. An improved biclustering algorithm and its application to gene expression spectrum analysis. Genom. Proteo. Bioinformatics. 2005;3:189–193. doi: 10.1016/S1672-0229(05)03024-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanay A, Sharan R, Shamir R. Discovering statistically significant biclusters in gene expression data. Bioinformatics. 2002;18(Suppl. 1):S136–S144. doi: 10.1093/bioinformatics/18.suppl_1.s136. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Kao MC, Wong WH. Transitive functional annotation by shortest-path analysis of gene expression data. Proc. Natl Acad. Sci. USA. 2002;99:12783–12788. doi: 10.1073/pnas.192159399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li KC. Genome-wide coexpression dynamics: theory and application. Proc. Natl Acad. Sci. USA. 2002;99:16875–16880. doi: 10.1073/pnas.252466999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 37.Reich M, et al. GeneCluster 2.0: an advanced toolset for bioarray analysis. Bioinformatics. 2004;20:1797–1798. doi: 10.1093/bioinformatics/bth138. [DOI] [PubMed] [Google Scholar]
- 38.Kapushesky M, et al. Expression Profiler: next generation–an online platform for analysis of microarray data. Nucleic Acids Res. 2004;32:W465–W470. doi: 10.1093/nar/gkh470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu CJ, Kasif S. GEMS: a web server for biclustering analysis of expression data. Nucleic Acids Res. 2005;33:W596–W599. doi: 10.1093/nar/gki469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Hoon MJ, et al. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin G, et al. Reproducibility Probability Score-incorporating measurement variability across laboratories for gene selection. Nat. Biotechnol. 2006;24:1476–1477. doi: 10.1038/nbt1206-1476. [DOI] [PubMed] [Google Scholar]
- 43.Wang QT, et al. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev. Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- 44.Pletcher SD, et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 2002;12:712–723. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 45.Lund J, et al. Transcriptional profile of aging in C. elegans. Curr. Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 46.Gilad Y, et al. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 2005;15:674–680. doi: 10.1101/gr.3335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilad Y, et al. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 48.Voolstra C, et al. Contrasting evolution of expression differences in the testis between species and subspecies of the house mouse. Genome Res. 2007;17:42–49. doi: 10.1101/gr.5683806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao BY, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol. Biol. Evol. 2006;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- 50.Sartor MA, et al. A new method to remove hybridization bias for interspecies comparison of global gene expression profiles uncovers an association between mRNA sequence divergence and differential gene expression in Xenopus. Nucleic Acids Res. 2006;34:185–200. doi: 10.1093/nar/gkj413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moehring AJ, Teeter KC, Noor MA. Genome-wide patterns of expression in Drosophila pure species and hybrid males. II. Examination of multiple-species hybridizations, platforms, and life cycle stages. Mol. Biol. Evol. 2007;24:137–145. doi: 10.1093/molbev/msl142. [DOI] [PubMed] [Google Scholar]
- 52.Jiao Y, et al. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraser HB, et al. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarroll SA, et al. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 55.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 56.Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 57.Wang DY, Kumar S, Hedges SB. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc. Biol. Sci. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benton MJ, Ayala FJ. Dating the tree of life. Science. 2003;300:1698–1700. doi: 10.1126/science.1077795. [DOI] [PubMed] [Google Scholar]
- 59.Kumar S, et al. Placing confidence limits on the molecular age of the human-chimpanzee divergence. Proc. Natl Acad. Sci. USA. 2005;102:18842–18847. doi: 10.1073/pnas.0509585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 61.Hekimi S, Guarente L. Genetics and the specificity of the aging process. Science. 2003;299:1351–1354. doi: 10.1126/science.1082358. [DOI] [PubMed] [Google Scholar]
- 62.Rodwell GE, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2:e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.St-Onge L, et al. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 64.Nishi M, et al. A case of novel de novo paired box gene 6 (PAX6) mutation with early-onset diabetes mellitus and aniridia. Diabet. Med. 2005;22:641–644. doi: 10.1111/j.1464-5491.2005.01469.x. [DOI] [PubMed] [Google Scholar]
- 65.Yasuda T, et al. PAX6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes. 2002;51:224–230. doi: 10.2337/diabetes.51.1.224. [DOI] [PubMed] [Google Scholar]
- 66.Grzeskowiak R, et al. Insulin responsiveness of the glucagon gene conferred by interactions between proximal promoter and more distal enhancer-like elements involving the paired-domain transcription factor Pax6. J. Biol. Chem. 2000;275:30037–30045. doi: 10.1074/jbc.M000984200. [DOI] [PubMed] [Google Scholar]
- 67.Scardigli R, et al. Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron. 2001;31:203–217. doi: 10.1016/s0896-6273(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 68.Richardson J, Cvekl A, Wistow G. Pax-6 is essential for lens-specific expression of zeta-crystallin. Proc. Natl Acad. Sci. USA. 1995;92:4676–4680. doi: 10.1073/pnas.92.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de la Monte SM, et al. Therapeutic rescue of neurodegeneration in experimental type 3 diabetes: relevance to Alzheimer's disease. J. Alzheimers Dis. 2006;10:89–109. doi: 10.3233/jad-2006-10113. [DOI] [PubMed] [Google Scholar]
- 70.Evans JL, et al. Oxidative stress and stress-activated signaling pathways: a unifying hypothesis of type 2 diabetes. Endocr. Rev. 2002;23:599–622. doi: 10.1210/er.2001-0039. [DOI] [PubMed] [Google Scholar]
- 71.Lemaire P, Livingstone DR. Aromatic hydrocarbon quinone-mediated reactive oxygen species production on hepatic microsomes of the flounder (Platichthys flesus L.) Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1997;117:131–139. doi: 10.1016/s0742-8413(97)00060-1. [DOI] [PubMed] [Google Scholar]
- 72.Bolton JL, et al. Role of quinones in toxicology. Chem, Res, Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 73.SantaCruz KS, et al. Regional NAD(P)H:quinone oxidoreductase activity in Alzheimer's disease. Neurobiol. Aging. 2004;25:63–69. doi: 10.1016/s0197-4580(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 74.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 75.Wickelgren I. Tracking insulin to the mind. Science. 1998;280:517–519. doi: 10.1126/science.280.5363.517. [DOI] [PubMed] [Google Scholar]
- 76.Marx J. Neurobiology. New clue to the cause of Alzheimer's. Science. 2001;292:1468. doi: 10.1126/science.292.5521.1468a. [DOI] [PubMed] [Google Scholar]
- 77.Taubes G. Neuroscience. Insulin insults may spur Alzheimer's disease. Science. 2003;301:40–41. doi: 10.1126/science.301.5629.40. [DOI] [PubMed] [Google Scholar]
- 78.Begum N, et al. Active Rho kinase (ROK-alpha) associates with insulin receptor substrate-1 and inhibits insulin signaling in vascular smooth muscle cells. J. Biol. Chem. 2002;277:6214–6222. doi: 10.1074/jbc.M110508200. [DOI] [PubMed] [Google Scholar]
- 79.Farah S, et al. A rho-associated protein kinase, ROKalpha, binds insulin receptor substrate-1 and modulates insulin signaling. J. Biol. Chem. 1998;273:4740–4746. doi: 10.1074/jbc.273.8.4740. [DOI] [PubMed] [Google Scholar]
- 80.Furukawa N, et al. Role of Rho-kinase in regulation of insulin action and glucose homeostasis. Cell. Metab. 2005;2:119–129. doi: 10.1016/j.cmet.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 81.Baudry A, et al. Genetic manipulation of insulin signaling, action and secretion in mice. Insights into glucose homeostasis and pathogenesis of type 2 diabetes. EMBO Rep. 2002;3:323–328. doi: 10.1093/embo-reports/kvf078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakamura Y, et al. Marked increase of insulin gene transcription by suppression of the Rho/Rho-kinase pathway. Biochem. Biophys. Res. Commun. 2006;350:68–73. doi: 10.1016/j.bbrc.2006.08.192. [DOI] [PubMed] [Google Scholar]
- 83.Homer S, Peinado M. On the Performance of Polynomial-time CLIQUE Approximation Algorithms on Very Large Graphs. Boston University Technical Report. 1994. http://www.cs.bu.edu/techreports/pdf/1994-001-maxclique.pdf.