Abstract
MolProbity is a general-purpose web server offering quality validation for 3D structures of proteins, nucleic acids and complexes. It provides detailed all-atom contact analysis of any steric problems within the molecules as well as updated dihedral-angle diagnostics, and it can calculate and display the H-bond and van der Waals contacts in the interfaces between components. An integral step in the process is the addition and full optimization of all hydrogen atoms, both polar and nonpolar. New analysis functions have been added for RNA, for interfaces, and for NMR ensembles. Additionally, both the web site and major component programs have been rewritten to improve speed, convenience, clarity and integration with other resources. MolProbity results are reported in multiple forms: as overall numeric scores, as lists or charts of local problems, as downloadable PDB and graphics files, and most notably as informative, manipulable 3D kinemage graphics shown online in the KiNG viewer. This service is available free to all users at http://molprobity.biochem.duke.edu.
INTRODUCTION
The atomic models of proteins and nucleic acids that come from X-ray crystallography and NMR are our most accurate sources of 3D information about these molecules, far more reliable than computed structures from modeling or simulation. They are best when determined at high resolution or with many restraints per residue, but even then are not perfect: nearly all structures in the Protein Data Bank (PDB; 1) have a few local errors, such as backwards-fit branched sidechains, flipped amides and imidazoles, incorrect sugar puckers, misoriented ligands, misidentified ‘waters’ and local errors in chain tracing. Such errors are usually due to misinterpretations of ambiguous experimental data. The ambiguity can often be resolved by considering additional information (such as steric interactions between atoms). This is the kind of structure validation data provided by MolProbity (2). The current paper reports on both protein and nucleic acid functionality in MolProbity, and on recent enhancements.
MolProbity is related to programs such as PROCHECK (3), PROCHECK-NMR (4), WHATIF (5) and OOPS (6), which provide both overall statistical evaluations and flags of local problem areas, concentrating primarily on geometrical measures that can be analyzed from the model. Other validation utilities analyze aspects of the model-to-data agreement, such as SFCHECK (7), real-space residuals (8), water-peak analysis in DDQ (9) and the now the almost universally utilized Rfree value (10) for X-ray, in addition to NMR R-factors (11) and RPF scores (12) for NMR. Whereas global validation measures serve the function of judging whether a structure meets accepted current practice, local measures are especially important to users of structures, since no level of global quality can guarantee protection against a large local error in the region of specific interest.
MolProbity is unique in offering all-atom contact analysis and up-to-date, high-accuracy Ramachandran and rotamer distributions. It is also broader in scope than many validation programs: it applies to both X-ray and NMR structures, and to both proteins and nucleic acids. Finally, it is useful to both ‘consumers’ and ‘producers’ of structural models: consumers can check that regions of interest are accurate, and producers can find and fix errors during fitting and refinement. MolProbity also focuses producers' efforts on the areas that actually need attention, thereby making accurate structure determination much faster. Experience has shown that most flagged problems are worth examining, and that most such errors can be corrected offline (13), either by traditional rebuilding methods or with our tools in Mage (14) and KiNG (15).
MATERIALS AND METHODS
MolProbity is implemented in PHP as a web server located at http://molprobity.biochem.duke.edu. It provides a graphical interface to a collection of Richardson lab programs for validation and structure correction. However, MolProbity is not a mere job-submission form; it is a complex web application that offers multiple modes of use, integrates many different kinds of information and suggests courses of action based on that information.
MolProbity uses a variety of physics- and knowledge-based algorithms to analyze a structure. The primary basis of its enhanced effectiveness is all-atom contact analysis, as implemented in Probe (16). All-atom contacts are exquisitely sensitive to a wide variety of local misfittings, but they are not yet available in other validation systems. They do require explicit hydrogen atoms, but MolProbity can add and optimize these using Reduce (17), while at the same time detecting and automatically fixing flipped Asn, Gln and His sidechains. MolProbity also uses carefully filtered, high-accuracy Ramachandran and rotamer distributions to check mainchain and sidechains for conformational outliers. Finally, it reports on some novel geometric indicators of misfitting, such as the Cβ deviation (18) and the base-phosphate perpendicular distance. The different types of analysis are synthesized into two integrated reports on the structural model: one tabular and one graphical.
Input
The primary input for MolProbity is structural models in PDB format. Models may be uploaded directly, or MolProbity can fetch them from the PDB or from the Nucleic Acid Database (NDB; 19), given the appropriate identifier. Models may come from crystallography, NMR or computation. However, some analyses are limited to single models rather than ensembles, and others focus on problems that are unique to structures determined by one particular method.
Additional data may optionally be uploaded to supplement the analysis. For instance, users may provide custom ‘het’ dictionaries to aid in placing hydrogens on any unusual small-molecule groups that their model may contain. Users can also upload electron density maps or have them fetched from the Electron Density Server (8) to view them overlaid with the 3D validation reports.
Kinemage files may also be uploaded into MolProbity and then viewed online in KiNG. This allows a user to view preexisting kinemage graphics without installing either KiNG or Mage on their local computer. Simple kinemages can also be constructed from a PDB file within MolProbity and then viewed online and/or downloaded.
Validation and analysis
Adding hydrogens
Explicit hydrogens are required for all-atom contact analysis, because they account for half of all atoms and three-quarters of all contacts in a typical biomolecule and the united-atom approach does not represent their interactions well. MolProbity adds hydrogens to files that do not already have them using the program Reduce (17); it can also redo and optimize placement of preexisting hydrogens. In brief, Reduce starts by placing hydrogens geometrically, and then analyzes each local H-bond network to optimize those H atoms that can move (e.g. rotatable hydroxyls) to avoid clashes and favor H-bonds. Most methyls are kept staggered, and only the first layer of water is considered [see (17) for details and rationale]. We have recently implemented a dynamic programming algorithm that greatly speeds up this process (see the Results section).
Flipping Asn/Gln/His
In the process of optimizing hydrogen positions, Reduce also searches for certain kinds of common fitting errors that may lead to suboptimal H placement. Specifically, in determining a structure by X-ray crystallography, it is easy to misorient the ends of Asn, Gln and His sidechains by 180° because the electron density is symmetric (except at extremely high resolutions). This type of misfitting changes both hydrogen-bonding and steric considerations (NH2 occupies more space than O). By default, Reduce explicitly tests both orientations for Asn, Gln and His sidechains while optimizing H placement, and makes a change if the score improvement exceeds a (settable) threshold. MolProbity illustrates the proposed corrections with 3D kinemage graphics and allows the user to veto changes before proceeding (needed only very rarely). MolProbity's assignments have been confirmed by independent experimental data (20). This Asn/Gln/His flip step can provide meaningful improvements (see the Discussion section); is very fast, easy, and reliable; and has become standard practice in many labs between rounds of crystallographic refinement.
All-atom contacts
Once hydrogens are present, all-atom contacts are calculated by the program Probe (16). Probe uses a rolling-probe algorithm to calculate colored dot surfaces. Blue dots appear when atoms approach within 0.5 Å, become green within 0.25 Å of touching, and yellow for slight overlaps. When nonbonded atom pairs overlap significantly, the contact dots are pale green for hydrogen bonding, but they become red spikes for disfavored (and impossible) clashes. Because van der Waals energy rises so quickly as atoms overlap, red spikes do not represent strained valid conformations; rather, they indicate inconsistencies in the model.
Probe analysis defines contact type and atom–atom gap or overlap at each dot, which can be integrated over each contact patch (default: 16 dots/Å2) to define various quantitative scores (16) for purposes such as automated corrections. However, we find the simple clashscore (number of overlaps > 0.4 Å per thousand atoms) is useful and well behaved as an overall validation metric. Many crystallographers make a point of trying to lower their clashscore as well as R and Rfree over the course of refining a structure.
Ramachandran and rotamer outliers
MolProbity uses quality-filtered, high-accuracy, empirical Ramachandran distributions (18) and sidechain rotamer distributions (21) for about 100,000 residues from our Top500 database to identify and score conformational outliers in protein mainchain and sidechain. We calculated explicit dihedral-angle distributions smoothed in the appropriate multi-dimensional space (not just conformer libraries), giving Ramachandran criteria for four classes of protein backbone (Gly, Pro, pre-Pro and the general case) and sidechain rotamer criteria for all 18 rotatable amino acids. We contour those distributions to classify as outliers the least probable 1% of sidechain conformations and the least probable 0.05% of general-case backbone conformations (0.2% for the smaller Gly, Pro and pre-Pro distributions). For the low B-factor residues in our database of high-quality structures, most of these rare conformations are real but strained. However, in a typical model with 5–10% of sidechain rotamers below the ‘outlier’ threshold, most of the outliers simply are mistakes and should be corrected, with only the few cases kept that are unambiguously constrained both by the experimental data and by their structural interactions.
It is possible to do a similar analysis for RNA backbone, since it also adopts rotameric conformations (22). With many more degrees of freedom and much less high-resolution data available, smoothed distributions in the 7-dimensional dihedral space are not feasible. However, conformer assignments and a rough quality index have been implemented, and will be included in MolProbity once a consensus list of conformers and their names have been officially defined by the RNA Ontology Consortium (23).
Cβ deviations
Cβ deviation measures a particularly significant kind of bond angle distortion in proteins; it is the distance from the modeled Cβ position to the expected ideal position calculated from the backbone coordinates (18). A large Cβ deviation (>0.25 Å) often signals an incompatibility between the sidechain and mainchain conformation; for instance, a sidechain fit 180° backwards. While the distortion can occur in various bond angles around Cα, a good refinement program often spreads the distortion around, so that no one angle is sufficiently bad to attract attention.
Sugar puckers
RNA sugar pucker (C3′ endo or C2′ endo) is strongly correlated to the perpendicular distance between the following (3′) phosphate and either the plane of the base or the C1′–N1/9 glycosidic bond vector. Incorrectly chosen sugar puckers also often result in out-of-range values for the epsilon dihedral. This is important information, because a sugar pucker is very difficult to determine directly from the electron density at resolutions typical for large RNAs. MolProbity checks epsilon angles and checks the modeled sugar pucker against the base-phosphate distance; it flags outliers as potentially having the wrong pucker.
Output
MolProbity produces several types of output:
A modified PDB-format file with optimized explicit hydrogens, corrected Asn/Gln/His flips, or both.
3D kinemage graphics that highlight local clusters of errors in the context of the structure. These may be viewed online, or downloaded and used for rebuilding in Mage (14) or KiNG (15).
Tabular summaries and lists of the same validation data. This now includes a ‘to-do list’ that can be read into the crystallographic rebuilding program Coot (24).
Analyses of molecular interface contacts.
Machine-readable (plain text) quantitative data underlying the above reports. This may be used as input for further analysis by outside tools.
User interface and automation
MolProbity is typically accessed by pointing a web browser to http://kinemage.biochem.duke.edu (where related software and documentation are available) and then clicking the MolProbity logo. The current interface has a main page that evolves during the session as the user makes choices. It typically displays a choice of structures to work on, a list of recommended and alternative actions for the selected structure, a ‘lab notebook’ summary of recent actions taken, a form for inputting additional files and a partial list of downloadable ‘result’ files. The left margin and bottom of the page have links to the full list of downloadable files, the full lab notebook, tutorials, help and other information.
Java is not required to use the site, but users with Java installed can see 3D kinemage molecular graphics and 3D validation reports directly in the browser via the KiNG applet.
While typical use is through a web browser, there are other ways to use MolProbity. For instance, structural genomics centers with many structures to analyze have written scripts to do automated batch processing on their own local computers. Thanks to the new MolProbity architecture, simple command-line PHP scripts can leverage all the power of the MolProbity website; several sample scripts are included with the source code. Also, several pharmaceutical companies concerned about data security have installed private MolProbity servers in-house, for both scripted and web use. Finally, individual component programs like Probe and Reduce can be downloaded and run independently for projects that need bulk runs or more complex options. Regardless, the code is free and open source for all, and can be downloaded from the MolProbity site or the Kinemage homepage.
Typical workflow
MolProbity is a flexible tool and can be used for many different purposes, but typical use follows one of two patterns, depending on whether the user is a ‘producer’ of structures or a ‘consumer’ of someone else's structures.
Producers typically upload a PDB file after a round of refinement, then add hydrogens while allowing Asn/Gln/His flips. (Unless this is the structure's first pass through MolProbity, there will probably be few or no flips.) They run the full suite of all-atom contacts and geometric analysis, and then either download the multi-criterion kinemage to fix problems in Mage or KiNG, or else work their way through the multi-criterion chart while fixing problems in a rebuilding program like O (6) or Coot (24), in either case starting from the flip-corrected PDB file (downloadable with or without hydrogens). After all the tractable problems are fixed, the structure is submitted for further refinement, and the cycle repeats.
On the other hand, consumers of structures generally fetch their models by PDB or NDB code. They add hydrogens and generally allow Asn/Gln/His flips; there will usually be several. (If flips occurred in the region of interest, it is important to use the flip-corrected PDB file for subsequent work.) They run the full suite of all-atom contacts and geometric analysis, and use the multi-criterion kinemage and/or chart to check for indicators of errors around specific sites of interest. They may run MolProbity on several related structure files, to help decide which one is most accurate or best suited to their purposes.
RESULTS
The MolProbity server has been operating continuously for more than 5 years now, with hundreds of different users per month and thousands of sessions. Thousands of different scientists have accessed it since its inception, and more structure files have been run through MolProbity than the number of files deposited in the PDB. About 80% of MolProbity sessions use uploaded files, and 20% fetch database files.
MolProbity has seen many improvements since its earlier published description (2): both the graphical and tabular reports have been consolidated into ‘multi-criterion’ evaluations; capabilities have been added for dealing with NMR ensembles and analyzing interface contacts; RNA functionality has been expanded; the interface has been redesigned for more power while remaining simple to use; the Reduce hydrogen optimization algorithm has been greatly sped up; and Reduce, KiNG and MolProbity have had significant rewrites of their internal code to be cleaner and more robust. These changes are described below.
Multi-criterion kinemages and charts
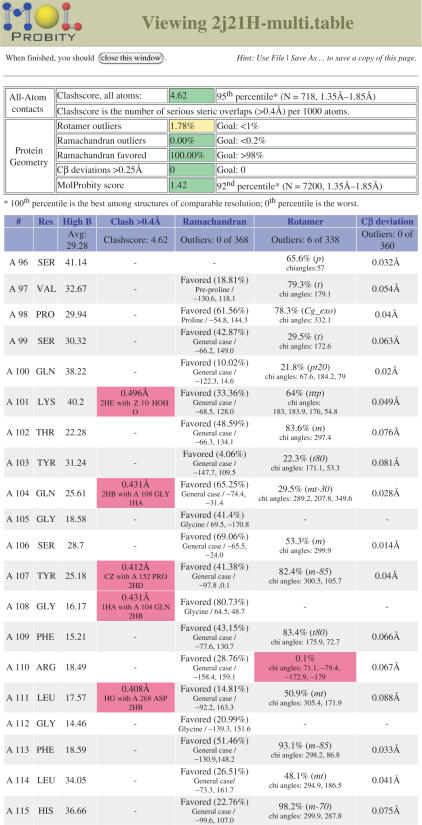
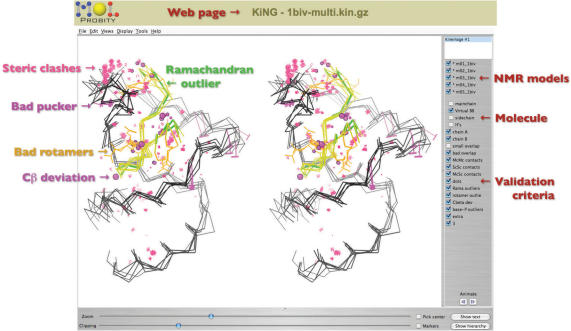
All the analyses that MolProbity performs are reported via two ‘multi-criterion’ displays. The multi-criterion chart provides a sortable, spreadsheet-like view of residue-by-residue statistics; outliers are flagged in hot pink for easy identification (see Figure 1). Any column can be sorted in the order of outlier severity. The multi-criterion kinemage shows the same information as color-coded glyphs superimposed on the 3D molecular structure (see Figure 2). This kinemage can be viewed directly in Java-enabled web browsers by way of the KiNG plugin. The two displays have complementary strengths: the chart can report numeric details, such as the highest crystallographic B-factor in the residue (reflecting local uncertainty) and the dihedral angles and probability of a rotamer outlier, while the kinemage highlights local clusters of problems involving residues that are not adjacent in sequence. (Such a cluster is typically caused by one underlying error, which can often be diagnosed visually.)
Figure 1.
Multi-criterion chart for 2J21, a crystal structure of the p53 DNA-binding core domain (37). The chart shows both overall statistics (top) and the first 20 residues of local data. Although a few steric clashes and one rotamer outlier are visible here (pink boxes) and might be worth trying to fix, this is an excellent structure overall; its resolution is 1.6Å, and compared to other structures at similar resolution, it ranks in the 92nd percentile for overall quality (MolProbity score).
Figure 2.
Multi-criterion kinemage of a 5-model NMR ensemble in side-by-side stereo, displayed in the KiNG applet. A small peptide (yellow) is bound to a short RNA hairpin (black), in file 1BIV (38). MolProbity has highlighted steric clashes (pink spikes), suspect RNA sugar puckers (magenta T's), outlier conformations of protein backbone (green) and sidechains (gold) and deviant bond angles around protein Cα's (magenta balls). On the right side, KiNG has controls for turning on or off the individual models, parts of the molecules (protein versus nucleic acid, Calphas versus full backbone, etc.) and validation criteria.
MolProbity also provides a summary of the different validation criteria, listing numbers of outliers and, in some cases, percentile ranking versus a PDB sample of structures at similar resolutions. We are currently exploring the possibility of summarizing further by creating a single number called the ‘MolProbity score’. Of course, no single number can capture all the information in a complete validation report. The final definition of the MolProbity score will be described in a future publication, but interested parties can try the current proposal (a weighted sum of clashes, Ramachandran not-favored and rotamer outliers) on the public server.
NMR ensemble analysis
Although the same basic types of analysis apply to NMR structures as X-ray structures, some differences must be taken into account. For instance, NMR models already have explicit hydrogens, and if their Asn/Gln/His sidechains are misplaced, there is no particular reason for them to be off by 180°. When it comes to reporting on the MolProbity analysis, single NMR models can be treated just like X-ray models. However, one generally wants to compare various members of the NMR ensemble. The sheer quantity of data (10–30 times or more) makes the chart format unwieldy, but the multi-criterion kinemage has been successfully redesigned to enable comprehensible and productive use for ensembles. As shown in Figure 2, there are separate controls for models and validation criteria, and one can easily view some or all models superimposed, or animate through them one at a time.
Molecular interface analysis
MolProbity now allows users to analyze the interfaces between specific sections of their structures in detail, using the all-atom contact methods from Probe. The user can customize which types of structure to look at (protein, nucleic acid, water, het, etc.) as well as which chains, for example, one can find the interface between chain A and all nucleic acid atoms within the structure, or perhaps all interactions between a het group and protein. The types of contacts — hydrogen bonds, van der Waals, etc. — may also be specified more narrowly if desired. In addition to a 3D kinemage illustrating contacts at the specified interface, a list is also produced for the contacting atom pairs, detailing the type of contact and its surface area (16 dots = 1 Å2). Such analysis comparing two different variant structures would show what interface changes occur upon introducing a different small molecule, or given a particular conformational change.
New user interface and architecture
The internals and externals of MolProbity have both been significantly rewritten since 2004. The outer face has remained centered around a single ‘main page’. To combat the added complexity of handling more than one structure, multiple NMR models and new analysis tools, the page is now more sensitive to context, displaying only the applicable options and even prioritizing them based on typical usage. It also features a ‘lab notebook’ that records what was done and the results, and a file browser with different kinds of results separated into folders.
The underlying architecture is significantly more robust, and presentation is more cleanly separated from logic. This opens the door to multiple user interfaces that efficiently reuse the same underlying functions: from a web browser, from the command line, via web services, etc. Obviously, the web and command-line interfaces already exist; a web services interface will likely be provided if there is demand.
The molecular graphics program KiNG was also significantly rewritten to improve its consistency and reliability. While outward changes are minimal, it is now easier to extend with custom plug-in modules, and its core can even be used as a standalone 3D graphics library in other programs.
Faster hydrogen optimization
The core algorithms underlying MolProbity have also been improved. The Snoeyink group at UNC has recently applied dynamic programming techniques to Reduce. Hydrogen placement optimization is a computationally challenging problem. A general formulation of the hydrogen-placement problem is NP-complete, requiring simultaneous optimization of the placement of rotatable hydroxyl hydrogens; lysine or N-terminal amines; methionine methyl or methylated base hydrogens (but not aliphatic methyls, which are relaxed and best modeled staggered); flippable Asn, Gln and His sidechains; His protonation; and all movable hydrogens on het groups. The formulation is identical to that of the sidechain-placement problem (25,26). Such complexity usually forces software to rely on stochastic techniques, approximation algorithms or brute force enumeration. However, because Reduce's scoring function is short ranged, the problem can generally be solved in polynomial time with dynamic programming.
The hydrogen placement problem may be formulated as a graph problem. In this graph, each hydroxyl, methyl or flippable group is represented as a vertex. Each conformation for such a group is represented as a vertex state. If and only if the scoring function for a pair (or triple, quadruple, etc.) of groups is non-zero for at least one assignment of conformations to the groups, then their vertices are connected by an edge (or a hyperedge of degree-3, degree-4, etc.). The complexity for a single problem instance depends on the connectivity of the graph — specifically, it is exponential in the treewidth of the graph (27–29). Because Reduce's scoring function defines low-treewidth problem instances, dynamic programming is able to rapidly and optimally assign conformations to groups (30).
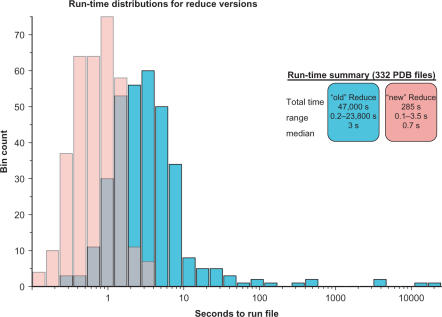
Most ‘cliques’ of interacting groups have six members or less, but the occasional larger cliques took hours to evaluate all possible states in the old Reduce; therefore, a limit was set to abort optimizing cliques with too many states (105 by default, 104 in MolProbity). Run with a limit of 107 states, the old Reduce's average time per file in a test set of 322 PDB files was 142 s and median time 3 s. The worst case was 6.6 h, and one clique could not be fully optimized (with 10 members and 1012 states, in 1OTF). With the new dynamic programming algorithm, the average time in the test set is down to 0.85 s (median 0.7 s), and the slowest case for the new algorithm took only 3.5 s. Furthermore, all files were fully optimized, because the algorithm no longer has to give up on large cliques. As illustrated in Figure 3, the speedup factor varies widely, but on average it is about 50-fold.
Figure 3.
Run-time distributions: improvement for the new version of Reduce. Optimizing hydrogens by exhaustive enumeration (cyan) was much slower than the new dynamic programming algorithm (pink). In many cases, this is the difference between a brief wait and completely infeasible calculation.
DISCUSSION
In practice, MolProbity has proved very useful during structure determination. Its success at improving structural accuracy is documented in Arendall (13); a test of 29 structural genomics crystal structures attained clash, rotamer and Ramachandran scores an order of magnitude better than the PDB average, resulting also in modest but meaningful improvements in traditional crystallographic critieria (0–4% drop in Rfree). Even though the changes affect relatively few atoms, the quality of the maps often improves noticeably throughout. General guidelines for using MolProbity in rebuilding and refinement are described by Richardson (14).
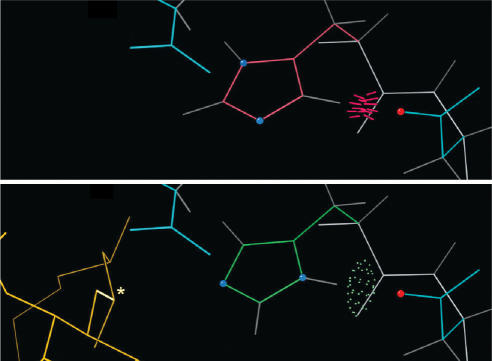
Figure 4 shows a specific example of how MolProbity is used during structure determination. The example comes from E. coli LpxA, an enzyme that catalyzes the first step in the biosynthesis of lipid A (31), as crystallized in complex with its product (32). The top panel shows His 125 as it was positioned in the original 1995 1LXA apo structure (33); that structure was used to phase the product complex by molecular replacement. MolProbity immediately suggested flipping His 125 based on a steric clash with Asp 126, thereby gaining an H-bond. His 125 had previously been proposed to function as a catalytic base (34); as the complex was refined, it became clear that His 125 was indeed part of the catalytic mechanism and did need to be flipped, in which position it could then interact with the substrate/product (bottom panel). Asp 126 was then also seen to provide an important backup interaction. This illustrates how quickly and easily MolProbity detects (and in some cases, fixes) local errors that can have a big impact on interpreting biochemical function.
Figure 4.
Example of a flipped active-site histidine found and corrected by Reduce. Top: His 125 of the apo LpxA clashes (red spikes) with nearby Asp 126 rather than H-bonding (green dots), prompting Reduce to suggest flipping it. Bottom: Once the product (gold) was modeled into its electron density (32), it was evident that the flipped position is necessary for His 125 to participate in catalysis.
For end users of structural data, MolProbity provides a thorough but easy way to choose critically among available structure files. For example, the p53 model evaluated in Figure 1 is not only at high resolution, but also scores in the 92nd percentile for structures at similar resolution, while other p53 structures range in overall quality all the way down to the 2nd percentile. However, MolProbity focuses more on local than on global validation, because most biological conclusions are based on the details of a few local regions. Even in a structure with excellent overall statistics, a cluster of validation outliers in the local region of interest is a cause for concern. Conversely, for crystal structures at least, a region free of validation flags is probably reliable even in a structure of modest overall quality. The bioinformatics study in Videau (35) illustrates the use of MolProbity's multiple criteria to differentiate a suspect motif sample from the reliable ones. Briefly, the known structures of type I DNA polymerases all share a local structural motif called a cis-Pro touch-turn — except for one structure that features a trans proline in that location. While the highest-resolution cis-Pro turn has good ϕ,ψ and bond angle values and no serious steric clashes, MolProbity showed that the trans-Pro turn contains a serious Ramachandran outlier and seven clashes with atomic overlap ≥0.4 Å. Thus, the authors concluded that the one exception was in fact an error, and that the cis-Pro touch-turn was strictly conserved through all of the type I DNA polymerases.
Although MolProbity is most complete for X-ray structures of proteins, it includes tools for working with both X-ray and NMR structures, and with both proteins and nucleic acids. Making this happen is a major challenge for any software with similarly broad aims: not only are there more kinds of data to deal with, but even familiar ones may have different properties. For example, it is sidechains for proteins but backbones for RNA that show conformations clustered into rotamers. Likewise, both NMR and X-ray models often contain steric clashes, but in X-ray they are highly constrained by the envelope of the electron density, whereas it is possible for NMR models to reduce clashes artificially by expanding slightly. One of the most persistently awkward differences is between single and multiple models. Although NMR ensembles are the most common source of multiple models, some crystallographers are advocating them as well (36). In either case, validation of ensembles hits information overload when reporting the results. As described above, we have been successful with ensemble multi-criterion kinemages, but it is still work for the future to design a manageable version of the multi-chart for large ensembles, to list both single-model validation flags and also show the relationships among models.
ACKNOWLEDGEMENTS
Maintenance and development of MolProbity is supported by NIH grant GM-073919. IWD acknowledges a Howard Hughes predoctoral fellowship, and JNB an NIH NRSA fellowship. We thank Ralf Grosse-Kunstleve for C++ advice, Scott Schmidler for help with scoring statistics, Mike Word for consultation on Reduce and Probe and Allison Williams for sharing her LpxA data. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM073919.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. ProCheck - A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 4.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 5.Vriend G. WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 6.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 7.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Cryst. D. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 8.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA. The Uppsala Electron-Density Server. Acta Cryst. D. 2004;60:2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 9.vandenAkker F, Hol WGJ. Difference density quality (DDQ): a method to assess the global and local correctness of macromolecular crystal structures. Acta Cryst. D. 1999;55:206–218. doi: 10.1107/S0907444998007161. [DOI] [PubMed] [Google Scholar]
- 10.Brunger AT. Free R-value - a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 11.Clore GM, Garrett DS. R factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc. 1999;121:9008–9012. [Google Scholar]
- 12.Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- 13.Arendall WB, Tempel W, Richardson JS, Zhou W, Wang S, Davis IW, Liu ZJ, Rose JP, Carson M, et al. A test of enhancing model accuracy in high-throughput crystallography. J. Struct. Func. Genomics. 2005;6:1–11. doi: 10.1007/s10969-005-3138-4. [DOI] [PubMed] [Google Scholar]
- 14.Richardson JS, Arendall WB, III, Richardson DC. New tools and data for improving structures, using all-atom contacts. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology, vol. 374. New York: Academic Press; 2003. pp. 385–412. [DOI] [PubMed] [Google Scholar]
- 15.Davis IW, Arendall WB, 3rd, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 2006;14:265–274. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, Richardson JS, Richardson DC. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogens. J. Mol. Biol. 1999a;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 17.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999b;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 18.Lovell SC, Davis IW, Arendall WB, III, Bakker PIWd, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins: Struct. Funct. Genet. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 19.Berman HM, Olson WK, Beveridge DL, Westbrook J, Gelbin A, Demeny T, Hsieh SH, Srinivasan AR, Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 1992;63:751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higman VA, Boyd J, Smith LJ, Redfield C. Asparagine and glutamine side-chain conformation in solution and crystal: a comparison for hen egg-white lysozyme using residual dipolar couplings. J. Biomol. NMR. 2004;30:327–346. doi: 10.1007/s10858-004-3218-y. [DOI] [PubMed] [Google Scholar]
- 21.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins: Structure, Function, and Genetics. 2000;40:389–408. [PubMed] [Google Scholar]
- 22.Murray LJ, Arendall WB, III, Richardson DC, Richardson JS. RNA backbone is rotameric. Proc. Natl. Acad. Sci. USA. 2003;100:13904–13909. doi: 10.1073/pnas.1835769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leontis NB, Altman RB, Berman HM, Brenner SE, Brown JW, Engelke DR, Harvey SC, Holbrook SR, Jossinet F, et al. The RNA Ontology Consortium: an open invitation to the RNA community. RNA. 2006;12:533–541. doi: 10.1261/rna.2343206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Pierce NA, Winfree E. Protein design is NP-hard. Prot. Engin. 2002;15:779–782. doi: 10.1093/protein/15.10.779. [DOI] [PubMed] [Google Scholar]
- 26.Leaver-Fay A, Kuhlman B, Snoeyink J. Pacific Symposium on Biocomputing, 2005. The Big Island, HI: World Scientific; 2005. An adaptive dynamic programming algorithm for the side chain placement problem; pp. 17–28. [PubMed] [Google Scholar]
- 27.Robertson N, Seymour PD. Graph Minors. I: Excluding a Forest. Journal of Combinatorial Theory Series B. 1983;35:39–61. [Google Scholar]
- 28.Arnborg S, Proskurowski A. Characterization and recognition of partial 3-trees. SIAM Journal of Algorithms and Discrete Methods. 1986;7:305–314. [Google Scholar]
- 29.Bodlaender HL. Proc. 15th Int. Colloq. Automata, Languages and Programming. Tampere, Finland: Springer, Lecture Notes in Computer Science 317; 1988. Dynamic programming on graphs with bounded treewidth; pp. 105–118. [Google Scholar]
- 30.Leaver-Fay A, Liu Y, Snoeyink J, Wang X. Faster placement of hydrogen atoms in protein structures by dynamic programming. Journal of Experimental Algorithms. 2007 In press. [Google Scholar]
- 31.Raetz CRH, Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 1990;265:1235–1238. [PubMed] [Google Scholar]
- 32.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A Modification Systems in Gram-Negative Bacteria. Ann. Rev. Biochem. 2007 doi: 10.1146/annurev.biochem.76.010307.145803. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raetz CRH, Roderick SL. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 34.Wyckoff TJ, Raetz CRH. The active site of Escherichia coli UDP-N-acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J. Biol. Chem. 1999;274:27047–27055. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 35.Videau LL, Arendall WB, III, Richardson JS. The cis-Pro touch-turn: a rare motif preferred at functional sites. Proteins: Struct. Funct. Bioinf. 2004;56:298–309. doi: 10.1002/prot.20101. [DOI] [PubMed] [Google Scholar]
- 36.Furnham N, Blundell TL, DePristo MA, Terwilliger TC. Is one solution good enough? Nat. Struct. Mol. Biol. 2006;13:184–185. doi: 10.1038/nsmb0306-184. discussion 185. [DOI] [PubMed] [Google Scholar]
- 37.Joerger AC, Ang HC, Fersht AR. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl. Acad. Sci. USA. 2006;103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye X, Kumar RA, Patel DJ. Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex. Chem. Biol. 1995;2:827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]