Abstract
Multiple Mapping Method with Multiple Templates (M4T) (http://www.fiserlab.org/servers/m4t) is a fully automated comparative protein structure modeling server. The novelty of M4T resides in two of its major modules, Multiple Templates (MT) and Multiple Mapping Method (MMM). The MT module of M4T selects and optimally combines the sequences of multiple template structures through an iterative clustering approach that takes into account the ‘unique’ contribution of each template, its sequence similarity to other template sequences and to the target sequences, and the quality of its experimental resolution. MMM module is a sequence-to-structure alignment method that is aimed at improving the alignment accuracy, especially at lower sequence identity levels. The current implementation of MMM takes inputs from three profile-to-profile-based alignment methods and iteratively compares and ranks alternatively aligned regions according to their fit in the structural environment of the template structure. The performance of M4T was benchmarked on CASP6 comparative modeling target sequences and on a larger independent test set and showed a favorable performance to current state-of-the-art methods.
INTRODUCTION
Comparative modeling is currently the most accurate protein structure prediction method (1). A prerequisite for successful comparative modeling is to find at least one suitable structure that shares a detectable sequence similarity spanning most of the modeled sequence (2). Accordingly, the two most critical steps in comparative modeling are: (i) identifying one or more templates, and (ii) calculating an accurate alignment between the target sequence and template structure(s) (3). The first step in comparative modeling is aided by several methods developed for fold-recognition (4–6) and profile-alignment (7,8) that allow an efficient recognition of remotely related sequences. Although these methods often identify more than one template structure, currently available modeling programs, and especially the automated servers, typically consider only one template for building a model for a target sequence. Meanwhile results at CASP meetings (9) and other reports(10,11) indicate that the use of multiple templates improves the quality of comparative models (10).
Accurate alignment of a target sequence to a template structure continues to be a bottleneck in producing good quality homology models. A number of alignment methods have been developed and are publicly available. However, none of these alignment methods consistently produces a better solution that is better than those from other methods (12,13). Furthermore, alignments produced by different methods are often better in some regions and worse in others when compared to one other. One possible solution to this problem is to consider several alignment methods and combine better-aligned parts into a unique solution (14).
The M4T server has been developed to address these issues by producing accurate alignments and models by minimizing the errors associated with the first two steps (template recognition and alignment) in comparative modeling. In the first step, protein structures are searched, compared and analyzed, and a number of candidates are selected to serve as templates. Next, to reduce errors associated with sequence-to-structure alignments, M4T uses an iterative implementation of the Multiple Mapping Method (MMM) (12) that considers solutions from several alignment methods and combines better-aligned parts into a unique solution, which, on average, is more accurate than any of the input alignments alone. In the final step, using these critical inputs, a default comparative protein structure model building is performed using Modeller (15).
M4T server
M4T server performs three main tasks in an automated manner (Figure 1): (i) template search and selection performed by the Multiple Template (MT) module; (ii) target sequence to template structure(s) alignment, performed by the Multiple Mapping Module (MMM) module (12) and (iii) model building, performed by Modeller (15).
Figure 1.
General overview of the algorithm: first, a PSI-BLAST search is performed with a query sequence, then template(s) are selected in the MT-module; subsequently, MMM-module performs sequence-to-structure alignment(s), and finally Modeller builds the protein model(s).
Template selection: MT module
The target sequence is used as query to search for homologous protein structure(s) that could serve as template(s) by running three iterations of PSI-BLAST (8) against PDB (16), with an E-value cutoff of 0.0001. Only those hits are selected where the sequence overlap with the target sequence is covering more than 60% of the actual SCOP domain length or more than 75% of the PDB chain length in case of a missing SCOP classification. After searching the PDB an iterative clustering procedure identifies the most suitable templates to combine, i.e. the least number of templates that can contribute the most to the model. Templates are selected or discarded according to a hierarchical selection procedure that accounts for sequence identity between templates and target sequence, sequence identity among templates, crystal resolution of the templates and contribution of templates to the target sequence (i.e. if a region is covered by several templates or by a single template only).
The result of the iterative clustering of templates is one or more groups of templates each containing one or more template structures (Figure 2). Within each cluster, all templates are aligned to the corresponding target sequence using the iterative-MMM approach (see later). In the last consolidation step the sequence-to-structure alignments of the overlapping clusters are combined. The overlapping parts are identified by first structurally superposing the templates, and subsequently by calculating an LGA_S score (17) on that superposition. If this score is greater than 70%, the overlapping clusters are combined using their alignment to the (same) target sequence as reference. If clusters of templates are not overlapping or the overlap between them is not sufficient for a structurally accurate superposition (i.e. less than six residues) then the target sequence is split into independent, separate parts and individual models are built for each ‘modelable’ part of the target sequence.
Figure 2.
Details of the MT and MMM modules of M4T. In the MT module the template candidates go through an iterative clustering and filtering process to select the least number of templates with a unique contribution to the target. The MMM module is an iterative implementation of the original Multiple Mapping Method using sequence profiles.
Target to template(s) alignment: MMM module
The target-to-template(s) alignments are calculated using an iterative implementation of the Multiple Mapping Method (12) (Figure 2). To construct profiles, the sequences of the target and template(s) are searched against the non-redundant database [NR (18)] of NCBI using five iterations of PSI-BLAST and with E-value cutoff of 0.0001. Next, BlastProfiler (19) is run to build representative sequence profiles for both the target and template sequences. BlastProfiler parses all iterations of PSIBLAST outputs, locates and stores those pairwise alignments between the query and database sequences that meet the filtering criteria. The values specified for filtering are: (i) lower and upper cutoffs for percent sequence identities between the hit and the query, as reported in the pairwise Blast alignment; default: 30 and 90%, respectively. (ii) Lower bound for alignment length; default: 30 residues. (iii) Maximal E-value for each hit; default: 0.0001. (iv) Minimal required coverage of the query in the alignment, in percentage; default: 30%. Typically, the PSI-BLAST output contains more than one alignment for the same hit sequence, especially when multiple iterations are performed. Such alternative alignments may include either the same or different regions of the hit sequence. Alignments to different regions of the target are kept as separate entries. Two alignments that involve the same hit sequence are considered redundant if the overlap is greater than 50%. Because alignments produced in later iterations contain more specific information about the sequence profile, these alignments are preferred over earlier ones in case of overlaps. The second major step in the selection of a set of representative hit sequences is to remove sequence redundancy using CD-HIT clustering program (20) at 40% identity level. Starting from the collected sequences, three separate profiles are calculated for each template(s) and target sequence, namely clustalw_d_profile, clustalw_m_profile and muscle_profile. The clustalw_d_profile and clustalw_m_profile are obtained using CLUSTALW (21) with default gap penalty function(clustalw_d_profile) and with modified gap penalty function (clustalw_m_profile). The CLUSTALW modified gap penalty function uses gap opening penalty of 5 and a gap extension penalty of 0.2, which are one-half of their corresponding default values and CLUSTALW was shown to perform competitively well with these parameters (12,19). The MUSCLE multiple sequence alignment program, with default parameters, is used to build muscle_profile (22). As a result, three separate profiles for the target sequence and three profiles for each template(s) are generated. Finally, the target profiles are aligned to the corresponding template profiles. At the end of this step, three alternative profile-to-profile-based sequence alignments are available, which are used as input to MMM (12).
Model building
Models are built with Modeller (15,23) using the default values for __model.top routine. Selected template(s) and optimized alignment(s) from the MT and MMM modules described earlier are provided as inputs.
Benchmarking model quality
Two measures are calculated to assess model quality. The DOPE score was published recently and it showed a favorable performance over other energy scores to rank models relatively to each other (24). DOPE score is useful if a user calculates several models for the same protein. In order to assess model quality in absolute terms we also calculate PROSA2003 scores and energy profile (25). The DOPE and PROSA2003 scores can be found in the header of the calculated coordinate file of the model while a separate html link leads to the PROSA2003 energy profile plot.
Performance of the method
The performance of M4T was extensively benchmarked on a set of 765 modeling cases and CASP targets, where a backdated version of PDB was used for searching for templates [to be published elsewhere; Fernandez-Fuentes, N., Rai, B., Madrid-Aliste, C., Fajardo, J. and Fiser, A. (2007) Comparative protein structure modeling by combining of multiple templates and optimizing sequence-to-structure alignments. Submitted].
All comparative model targets from CASP6 were tested by building models with M4T using the single best identified template and then by using multiple templates. In this setup we used the MMM alignment module of M4T to generate input alignments for both cases. For 11 out of 24 CASP6 comparative modeling targets it was possible to combine multiple templates. For all cases but one (T0269) the use of multiple templates provides a superior model in terms of RMSD and GDT_TS scores than the one based on a single best template. The most impressive improvement takes place in case of target sequence T0275 where the GDT_TS score increases from 55.37 to 72.41 when multiple templates are combined.
M4T also compared well with state-of-the art methods and human experts in protein modeling. M4T was compared with the single best models submitted to CASP6 by any group. It is less trivial to compare these results because alignments may be different due to different methods used, different profiles employed or manual editing. Also, certain users may have used information on multiple structures. In addition, expert users may have attempted side chain and loop modeling in certain parts of the models. An ultimate goal of automated structure prediction is to deliver models with a competitive accuracy to the ones created by ‘expert users’, and to do it in a fully automated way and in a short time. In 9 out of 24 cases, M4T outperformed the single best model submitted to CASP. As another qualitative comparison, in nine cases the differences between the best CASP model and M4T were too small to draw any conclusion, while in five and nine cases M4T or CASP models were significantly more accurate (for one case M4T did not return a model). Out of the 24 best CASP targets the largest population of targets that belonged to the same research group was 9, the second largest was 2. In this simplified comparison M4T would fare as the second best individual performer with 5 of the 24 best targets. While it is true that from a small number of test cases, such as at CASP, it is hard to conclude statistical significance (26) we perceive this performance as encouraging and a sign that automated methods are becoming competitive with the best expert users.
The real benefit of using multiple templates with Multiple Mapping Method is to generate more accurate models at low sequence identity levels (below 50%). As sequence identity decreases, the accuracy of models (in terms of RMSD to the experimental solution structure) that are built using multiple templates is better than the accuracy of models built using any single template alone as tested on random 765 modeling cases (Figure 3). In addition, on average, the length of the modeled sequence is longer when using multiple templates than when using a single template. When using multiple templates the length of model coverage increases by at least 1, 5, 10, 20 residues in 56, 21, 12.5, 2,5% of the cases, respectively, and the coverage is the same as in case of using single templates in 44% of the cases.
Figure 3.
RMSD (model compared to the actual experimental structure) versus sequence identity. Using a dataset of 765 proteins with known structures two sets of models were built: (1) using one template only (best E-value hit; light bars), (2) using multiple templates selected by MT (grey bars). The percentage of sequence identity is calculated between the hit sequence with the highest E-value and the query sequence. Error of the mean is indicated.
Design, implementation and use
M4T server is implemented on an Apache server running Fedora Core 5 operating system. The server is interfaced with a CGI Perl and Javascript coded web interface. The MT and MMM modules are coded in Perl and C++ language, respectively. Databases required by the server, namely, PDB (16) and NR (18), are locally installed and weekly updated. All the queries are submitted to a queuing system. Results are either displayed in HTML format or sent to the user by e-mail as a hyperlink.
Submitting a query
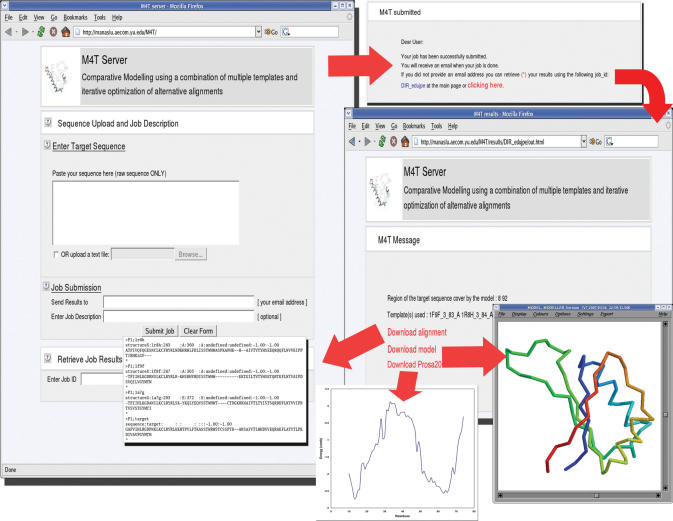
The M4T server has a straightforward interface (Figure 4). In order to use this server, the user must provide a target sequence, which can be entered in a text box, or can be uploaded as a text file. The target sequence must be in raw text containing one-letter amino acid codes (without any headers). Users may add a description of the sequence at the ‘Job Description’ field. If an e-mail address is provided the user is also notified by e-mail when the prediction is finished including a hyperlink where the results can be accessed. M4T assigns a unique job identifier for each submitted query (e.g. DIR_cA8r0n). This job identifier can be used to check the status of the submission (i.e. in queue, running, finished) and to retrieve the results by typing it in the ‘Job ID’ field at the submission page.
Figure 4.
Screenshots of the submission and results web pages.
Retrieving results
M4T returns a full atom model(s) in PDB format and the alignment(s) used to build the model. When the prediction process is finished, the server will send a notification by e-mail to the user (if an e-mail address was provided). Otherwise, users have to visit the submission page and access the results page by using the job identifier. Results are kept on the server for 5 days only.
Possible bottlenecks
Occasionally, M4T may fail to provide a prediction. The main reason is usually that PSI-BLAST (8) fails to find homologous protein structure(s) to the sequence. But even if PSI-BLAST succeeds to detect possible template(s), after running the MT module none of the PDB hits might be found to be suitable to model the target sequence. All details of the process are registered in a log file that users can examine. In addition users can contact the authors via e-mail to m4t@fiserlab.org for further information.
SUMMARY
A web server for comparative protein structure prediction is described that takes advantage of a recently developed new sequence to structure alignment technique and the optimal selection and use of multiple template structures. The most time-consuming parts of the M4T algorithm are the database searches and calculation of profiles (clustering). If there is no other competing job in the queue system the prediction typically is done in 5–20 min. The server is designed to deliver high quality comparative models to the non-experts users, with competitive quality to those produced by manual expert modelers.
ACKNOWLEDGEMENTS
This work was supported by NIH GM62519-04. Funding to pay the Open Access publication charges for this article was provided by NIH GM62519-04.
Conflict of interest statement. None declared.
REFERENCES
- 1.Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 2.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu.Rev.Biophys. Biomol. Struct. 2000;29:291. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 3.Fiser A. Protein structure modeling in the proteomics era. Expert Rev. Proteomics. 2004;1:97–110. doi: 10.1586/14789450.1.1.97. [DOI] [PubMed] [Google Scholar]
- 4.Domingues FS, Koppensteiner WA, Jaritz M, Prlic A, Weichenberger C, Wiederstein M, Floeckner H, Lackner P, Sippl MJ. Sustained performance of knowledge-based potentials in fold recognition. Proteins. 1999;37:112. [PubMed] [Google Scholar]
- 5.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 6.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 7.Li W, Pio F, Pawlowski K, Godzik A. Saturated BLAST: an automated multiple intermediate sequence search used to detect distant homology. Bioinformatics. 2000;16:1105. doi: 10.1093/bioinformatics/16.12.1105. [DOI] [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venclovas C, Zemla A, Fidelis K, Moult J. Assessment of progress over the CASP experiments. Proteins. 2003;53(Suppl 6):585. doi: 10.1002/prot.10530. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez R, Sali A. Evaluation of comparative protein structure modeling by MODELLER-3. Proteins. 1997;(Suppl. 1):50. doi: 10.1002/(sici)1097-0134(1997)1+<50::aid-prot8>3.3.co;2-w. [DOI] [PubMed] [Google Scholar]
- 11.Contreras-Moreira B, Fitzjohn PW, Bates PA. In silico protein recombination: enhancing template and sequence alignment selection for comparative protein modelling. J. Mol. Biol. 2003;328:593. doi: 10.1016/s0022-2836(03)00309-7. [DOI] [PubMed] [Google Scholar]
- 12.Rai BK, Fiser A. Multiple mapping method: a novel approach to the sequence-to-structure alignment problem in comparative protein structure modeling. Proteins. 2006;63:644–661. doi: 10.1002/prot.20835. [DOI] [PubMed] [Google Scholar]
- 13.Prasad JC, Comeau SR, Vajda S, Camacho CJ. Consensus alignment for reliable framework prediction in homology modeling. Bioinformatics. 2003;19:1682. doi: 10.1093/bioinformatics/btg211. [DOI] [PubMed] [Google Scholar]
- 14.Kosinski J, Gajda MJ, Cymerman IA, Kurowski MA, Pawlowski M, Boniecki M, Obarska A, Papaj G, Sroczynska-Obuchowicz P, et al. FRankenstein becomes a cyborg: the automatic recombination and realignment of fold recognition models in CASP6. Proteins. 2005;61(Suppl 7):106–113. doi: 10.1002/prot.20726. [DOI] [PubMed] [Google Scholar]
- 15.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 16.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rai BK, Madrid-Aliste CJ, Fajardo JE, Fiser A. MMM: a sequence-to-structure alignment protocol. Bioinformatics. 2006;22:2691–2692. doi: 10.1093/bioinformatics/btl449. Epub 2006 Aug 2623. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 24.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 26.Marti-Renom MA, Madhusudhan MS, Fiser A, Rost B, Sali A. Reliability of assessment of protein structure prediction methods. Structure (Camb.) 2002;10:435. doi: 10.1016/s0969-2126(02)00731-1. [DOI] [PubMed] [Google Scholar]