Abstract
All tools in the DAVID Bioinformatics Resources aim to provide functional interpretation of large lists of genes derived from genomic studies. The newly updated DAVID Bioinformatics Resources consists of the DAVID Knowledgebase and five integrated, web-based functional annotation tool suites: the DAVID Gene Functional Classification Tool, the DAVID Functional Annotation Tool, the DAVID Gene ID Conversion Tool, the DAVID Gene Name Viewer and the DAVID NIAID Pathogen Genome Browser. The expanded DAVID Knowledgebase now integrates almost all major and well-known public bioinformatics resources centralized by the DAVID Gene Concept, a single-linkage method to agglomerate tens of millions of diverse gene/protein identifiers and annotation terms from a variety of public bioinformatics databases. For any uploaded gene list, the DAVID Resources now provides not only the typical gene-term enrichment analysis, but also new tools and functions that allow users to condense large gene lists into gene functional groups, convert between gene/protein identifiers, visualize many-genes-to-many-terms relationships, cluster redundant and heterogeneous terms into groups, search for interesting and related genes or terms, dynamically view genes from their lists on bio-pathways and more. With DAVID (http://david.niaid.nih.gov), investigators gain more power to interpret the biological mechanisms associated with large gene lists.
INTRODUCTION
In the post-genomic era, biological interpretation of large gene lists derived from high-throughput experiments, such as genes from microarray experiments, is a challenging task. The first version of DAVID (the Database for Annotation, Visualization and Integration Discovery), released in 2003 (1,2), as well as a number of other similar publicly available high-throughput functional annotation tools (3–23), partially address the challenge by systematically mapping a large number of interesting genes in a list to associated Gene Ontology (GO) terms (10), and then statistically highlighting the most over-represented (enriched) GO terms out of a list of hundreds or thousands of terms. This increases the likelihood that the investigator will identify the biological processes most pertinent to the biological phenomena under study (19). While this tool is extremely useful and has been cited in hundreds of publications during the past three years, the development of other effective data mining algorithms, as additional components to the DAVID Bioinformatics Resources, will improve the power of investigators to analyze their gene lists from different biological angles. The newly added contents, functions and tool suites in the DAVID Bioinformatics Resources intend to address several issues that other tools have not been able to extensively address: (i) to dramatically expand the biological information coverage in the DAVID Knowledgebase by comprehensively integrating more than 20 types of major gene/protein identifiers and more than 40 well-known functional annotation categories from dozens of public databases; (ii) to address the enriched and redundant relationships among many-genes-to-many-terms (i.e. one gene could associate with many different, redundant terms and one term could associate with many genes) by developing a set of novel algorithms, such as the DAVID Gene Functional Classification Tool, the Functional Annotation Clustering Tool, the Linear Searching Tool, the Fuzzy Gene-Term Heat Map Viewer, etc.; (iii) to dynamically visualize genes from a users list within the most relevant KEGG and BioCarta pathways with the DAVID Pathway Viewer; (iv) to allow users to create and use customized gene backgrounds for typical gene-term enrichment analysis utilizing the improved computational power and (v) to facilitate efficient communication and experience exchange within the scientific community by moderating the DAVID Forum.
This article summarizes the key DAVID components and tool suites in the newly released DAVID Bioinformatics Resources, highlighting new or expanded analytic features that provide investigators with additional means to explore and extract biological meaning from large gene lists that users input to the system (Supplementary File 1). For in-depth algorithm information, appropriate references and supplementary materials are provided.
FEATURES AND FUNCTIONALITIES
Computational Infrastructure
The aim of the DAVID software design is to provide users with the simplest usability and fastest exploration speed through better internal software engineering practices. Therefore, the DAVID Bioinformatics Tools, as web-based applications on a Tomcat web server in a Linux machine (4-CPU for 3.5 GHz speed, 8 GB memory), requires no configuration and installation in the client's computers. Java is the primary language used for all of the server side components of the calculation engines and the Java Server Page (JSP) web interfaces, in a full object-oriented fashion. In-memory Java data objects holding all genes-to-annotation information up to 2.5 GB in size were developed to greatly increase the data IO speed compared to that through typical relational databases (e.g. Oracle). The Java Remote Method Invocation (RMI), a distributed computing technique, is also used to take advantage of multiple computing resources. A set of automated programs monitors many aspects of the web services in order to maximize the performance and minimize the down time period.
DAVID Knowledgebase
A highly integrated gene-annotation database with comprehensive data coverage is essential for the success of any high-throughput annotation algorithms. Due to the complex and distributed nature of biological research, our current biological knowledge is distributed among many redundant annotation databases maintained by independent groups. One gene could have several different identifiers within one or more database(s). Similarly, the biological terms associated with different gene identifiers for the same gene could be collected in different levels across different databases. Due to these issues, most high-throughput annotation tools rely on one, or at most a few, resource(s), which limits the analytic comprehensiveness and the level of throughput. The DAVID Knowledgebase is now built around the ‘DAVID Gene Concept’, a single linkage method to agglomerate tens of millions of gene/protein identifiers from a variety of public genomic resources (Table 1), including NCBI, PIR and UniProt (24–27), into broader secondary gene clusters, called the DAVID Gene Concept (Figure 1, and more technical details at http://david.abcc.ncifcrf.gov/helps/knowledgebase/DAVID_gene.html), Grouping these gene identifiers improves cross-referencing capability, allowing more than 40 categories of publicly available functional annotation to be comprehensively assigned to and centralized by the DAVID Gene Concept (Table 2, see Supplementary File 2 for a complete list of annotation sources and more technical details at http://david.abcc.ncifcrf.gov/helps/knowledgebase/DAVID_gene.html). To the best of our knowledge, this annotation coverage far exceeds that of the original DAVID database and those currently used by other similar high-throughput annotation tools. The DAVID knowledgebase not only increases the accessibility to a wide range of heterogeneous annotation data in one centralized location, but also enhances the comprehensiveness of high-throughput gene functional analysis by overlapping multiple biological aspects together. It also provides a solid foundation for the further development of more advanced high throughput analytic algorithms that may be added to the DAVID Bioinformatics Resources. More importantly, the entire DAVID Knowledgebase, in simple pair-wise text format files containing a broad, highly integrated annotation data collection, is freely available to the public (http://david.abcc.ncifcrf.gov/knowledgebase), which will benefit various high-throughput data mining projects by other research groups. The DAVID Knowledgebase is expected to be updated more frequently in the near future than its current annual update.
Table 1.
Over 22 types of gene identifiers integrated by the DAVID Gene Concept within the DAVID Knowledgebase
| Gene ID Type | Total ID | Unique Cluster |
|---|---|---|
| AFFY_ID | 2254679 | 845117 |
| ENTREZ_GENE_ID | 1734858 | 1602339 |
| GENPEPT_ACCESSION | 4065385 | 2511637 |
| GENBANK_ACCESSION | 16828735 | 2409120 |
| GENEBANK_ID | 20291282 | 2358084 |
| PIR_ACCESSION | 282281 | 258079 |
| PIR_ID | 308092 | 266645 |
| PIR_NREF_ID | 3355759 | 2677404 |
| REFSEQ_GENOMIC | 1866800 | 1552597 |
| REFSEQ_MRNA | 645831 | 561447 |
| REFSEQ_PROTEIN | 1644632 | 1373467 |
| REFSEQ_RNA | 1364 | 852 |
| UNIGENE | 161138 | 158938 |
| UNIPROT_ACCESSION | 2864344 | 2097488 |
| UNIPROT_ID | 2789453 | 2096712 |
| UNIREF100_ID | 2552342 | 2088692 |
| OFFICIAL_GENE_SYMBOL | 1693151 | 1600906 |
| FLYBASE_ID | 27109 | 26642 |
| HAMAP_ID | 63925 | 63822 |
| HSSP_ID | 265000 | 258750 |
| TIGR_ID | 120117 | 111699 |
| WORMBASE_ID | 43675 | 21243 |
| RGD_ID | 25230 | 25060 |
| NOT SURE | ALL IDs |
Any of the gene identifier types above can be cross-mapped to the DAVID Knowledgebase. ‘Not Sure’ is a new ID type specifically designed for the DAVID web site. For a given ‘not sure’ ID, all possible matching IDs will be systematically scanned across the entire DAVID collection.
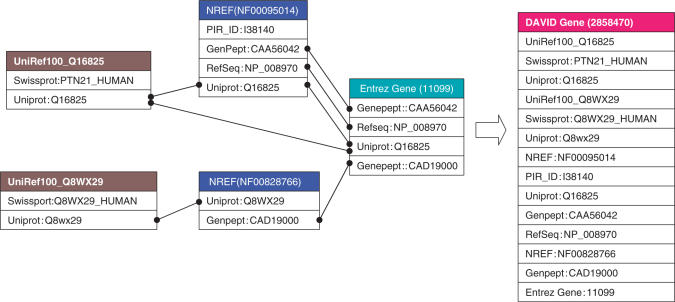
Figure 1.
A DAVID gene constructed by a single linkage algorithm. Two UniRef100 clusters, two NRef 100 clusters and one Entrez Gene cluster were systematically found sharing one or more protein identifiers with each other. The single-linkage rule can further iteratively agglomerate them as a whole into one DAVID gene. Thus, for this particular example of tyrosine-protein phosphatase non-receptor type 21 (PTPN21), the resulting DAVID gene is able to collect and integrate all gene/protein identifiers more comprehensively than each original gene cluster.
Table 2.
The wide-range collection of heterogeneous functional annotations in the DAVID Knowledgebase
| Ontology (>40 million records) | Protein Domain/Family (>15 millions) | Sequence Features (>21 millions) |
| GO_BIOLOGICAL PROCESS | BLOCKS_ID | ALIAS_GENE_SYMBOL |
| GO_MOLECULAR FUNCTION | COG_KOG_NAME | CHROMOSOME |
| GO_CELLULAR COMPONENT | INTERPRO_NAME | CYTOBAND |
| PANTHER_BIOLOGICAL PROCESS | PDB_ID | GENE_NAME |
| PANTHER_MOLECULAR FUNCTION | PFAM_NAME | GENE_SYMBOL |
| COG_KOG_ONTOLOGY | PIR_ALN | HOMOLOGOUS_GENE |
| P-P Interaction (>4 millions) | PIR_HOMOLOGY_DOMAIN | ENTREZ_GENE_SUMMARY |
| BIND | PIR_SUPERFAMILY_NAME | OMIM_ID |
| DIP | PRINTS_NAME | PIR_SUMMARY |
| MINT | PRODOM_NAME | PROTEIN_MW |
| NCICB_CAPATHWAY | PROSITE_NAME | REFSEQ_PRODUCT |
| TRANSFAC_ID | SCOP_ID | SEQUENCE_LENGTH |
| HIV_INTERACTION | SMART_NAME | SP_COMMENT |
| HIV_INTERACTION_CATEGORY | TIGRFAMS_NAME | Functional Category (>6.9 millions) |
| HPRD_INTERACTION | PANTHER_SUBFAMILY | PIR_SEQ_FEATURE |
| REACTOME_INTERACTION | PANTHER_FAMILY | SP_COMMENT_TYPE |
| Disease Association (∼9,000) | Pathways (>50 000) | SP_PIR_KEYWORDS |
| GENETIC_ASSOCIATION_DB | BioCarta | UP_SEQ_FEATURE |
| OMIM_DISEASE | KEGG_PATHWAY | Gene Tissue Expression (>1.0 million) |
| Literature (>2.8 millions) | PANTHER_PATHWAY | GNF Microarray |
| GENERIF_SUMMARY | PID | UNIGENE EST |
| PUBMED_ID | BBID | CGAP SAGE |
| HIV_INTERACTION_PUBMED_ID | KEGG_REACTION | CGAP EST |
Over 60 functional categories from dozens of independent public sources (databases) (see Supplementary File 2 for a complete list) are collected and integrated in the DAVID Knowledgebase.
DAVID Functional Annotation Tool Suite
This tool suite (http://david.abcc.ncifcrf.gov/summary.jsp), introduced in the first version of DAVID, mainly provides typical batch annotation and gene-GO term enrichment analysis to highlight the most relevant GO terms associated with a given gene list (2). The new version of the tool keeps the same enrichment analytic algorithm but with extended annotation content coverage, increasing from only GO in the original version of DAVID to currently over 40 annotation categories, including GO terms, protein–protein interactions, protein functional domains, disease associations, bio-pathways, sequence general features, homologies, gene functional summaries, gene tissue expressions, literatures, etc. (Table 2). The improved annotation coverage alone provides investigators with much more power to analyze their genes using many different biological aspects in a single space. Flexible options are provided to display results in an individual annotation chart report or a combined chart report. In addition to pre-built gene population backgrounds (e.g. Affy U133) used in gene-annotation enrichment analysis, with its improved computational power, the new tool accepts user-defined population gene list, an option rarely found in other similar web-based, high-throughput annotation tools. This feature was added in order to more specifically meet the users’ requirements for the best analytical results.
The DAVID Functional Annotation Clustering is a newly added feature (manuscript submitted, and more details at http://david.abcc.ncifcrf.gov/manuscripts/fuzzy_cluster/) to the DAVID Functional Annotation Tool. This function uses a novel algorithm to measure relationships among the annotation terms based on the degrees of their co-association genes to group the similar, redundant and heterogeneous annotation contents from the same or different resources into annotation groups. This reduces the burden of associating similar redundant terms and makes the biological interpretation more focused in a group level (Figure 2). The tool also provides a look at the internal relationships among the clustered terms. The clustered format is able to give a more insightful view about the relationships of annotations compared to the traditional un-clustered term report, over which similar annotation terms may be spread among hundreds, if not thousands, of other terms. In addition, to take full advantage of the well-known KEGG and BioCarta pathways, the new DAVID Pathway Viewer, another feature of the DAVID Functional Annotation Tool, can display genes from a user's list on pathway maps to facilitate biological interpretation in a network context.
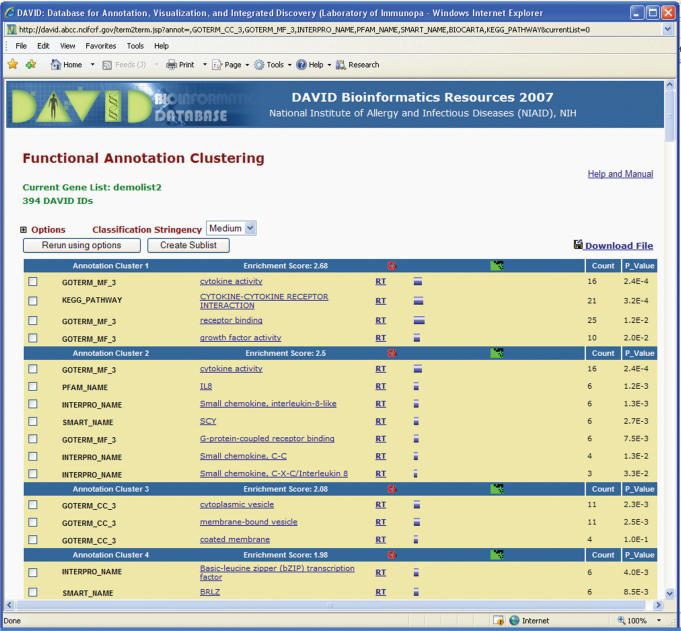
Figure 2.
An HTML report from the Functional Annotation Clustering. The annotation cluster 1 in the example shows that GO term cytokine activity, KEGG pathway cytokine–cytokine receptor interaction, and GO term receptor binding, etc. are grouped together. Thus, the different biological aspects regarding a relevant biology can be explored at the same time.
DAVID Gene Functional Classification Tool Suite
The DAVID Gene Functional Classification Tool (http://david.abcc.ncifcrf.gov/gene2gene.jsp) is a completely new component in the DAVID Bioinformatics Resources. The tool provides a novel way to functionally analyze a large number of genes in a high-throughput fashion by classifying them into gene groups based on their annotation term co-occurrence. This is accomplished and visualized by a set of new fuzzy classification algorithms, including a kappa statistics measurement of gene–gene functional relationship, a fuzzy multi-linkage partitioning method and a fuzzy genes-terms heat map visualization, etc. (manuscript submitted, and more details at http://david.abcc.ncifcrf.gov/manuscripts/fuzzy_cluster/). The power of the tool is that it allows users to simultaneously view the rich and redundant internal relationship of functionally related genes and their annotation terms within biological modules. Investigators are able to functionally analyze their gene list in a highly related many-genes-to-many-terms network context instead of a one-term-to-many-genes or a one-gene-to-many-terms view in the typical gene-annotation enrichment analysis.
DAVID Gene ID Conversion Tool Suite
A significant number of different types of gene/protein identifiers, not mutually mapped to each other across three independent resources, NCBI, PIR and UniProt (25,26,28), are now maximally integrated in the DAVID Knowledgebase (Figure 1, more details at http://david.abcc.ncifcrf.gov/helps/knowledgebase/DAVID_gene.html), whose scope is more expansive than one system only. Even though the DAVID Knowledgebase is used primarily for improvement of annotation terms integration and coverage, such comprehensive gene identifier coverage and cross-referencing capability could itself be very useful for researchers to convert their gene/protein identifiers from one type to another among over 20 major types of identifier systems (Table 1). Thus, with the newly introduced DAVID Gene ID Conversion Tool (http://david.abcc.ncifcrf.gov/conversion.jsp), interesting genes derived from one identifier system can be quickly translated to other gene identifier types preferred by a given annotation resource. In addition, the DAVID Gene ID Conversion Tool provides a ‘not sure’ type for ambiguous gene identifiers, whereby the tool can systematically suggest the potential type(s). For instance, a user has a gene ID ‘3558’ without ID type information. DAVID Gene ID Conversion Tool will scan all possibilities across all gene ID systems collected in the DAVID Knowledgebase. Two choices will be suggested, i.e. ‘3558’ could be an Entrez Gene ID for IL2 (human) or a Genbank ID for CNA1 (yeast). Thus, the user can make a decision based on above information.
DAVID Gene Name Batch Viewer
After obtaining a list of interesting genes, probably the first question researchers will ask is ‘What are the names of my genes?’ Even though it is a simple question, most high-throughput annotation tools do not answer it in a straightforward way. The new DAVID Gene Name Batch Viewer is designed to simply list the gene names for all given genes. In addition, hyperlinks are provided on each gene entry, allowing users to explore in depth other functional information around the gene. Thus, this tool provides users with a first glance and initial ideas about their interesting genes before proceeding to analysis by other more comprehensive analytic tool. Moreover, hyperlinks, labeled as ‘RT’, are provided for each gene in order to search other functionally related genes in user's gene list or the entire genome. The search is based on co-occurrence of annotations between genes (more details at http://david.abcc.ncifcrf.gov/helps/linear_search.html).
DAVID NIAID Pathogen Browser
The National Institute of Allergy and Infectious Diseases (NIAID) has defined category A, B and C priority pathogens (http://www3.niaid.nih.gov/Biodefense/bandc_priority.htm), which have subsequently become important in biodefense research funding, attracting broad interest from the research community. Since the organisms listed in these categories may not be familiar to researchers who have recently joined the emerging field, the DAVID NIAID Pathogen Browser (http://david.abcc.ncifcrf.gov/GB.jsp) is provided as a quick starting point for them to search the most relevant genes in the organisms by biological key words of interests. A large list of genes retrieved from the search could be further transferred to the DAVID Bioinformatics Resources for in-depth functional analysis with any of the previously mentioned tools. Although the tool is still in its early stage, it may help researchers gain understanding of the genes related to a priority pathogen of interest. More development is ongoing to extend the searching scope to all available genomes and annotations collected in DAVID knowledgebase.
DAVID API Services
DAVID API services (http://david.abcc.ncifcrf.gov/api/) are newly added features that allow users to directly pass gene list to various DAVID tools via a set of pre-defined URLs instead of DAVID submission forms. Thus, DAVID tools can easily serve as part of the analytic pipeline in other bioinformatics web sites. They can also be used in bioinformatics scripts to automate functional annotation for large number of gene lists, which are too many to be accomplished by the manual procedures.
CONCLUSION
The newly released DAVID Bioinformatics Resources are an expanded version of the original DAVID. It provides a set of powerful, novel tools that researchers can use to explore their large gene lists in depth from many different biological angles (Figure 3) in order to extract associated biological meanings to the greatest extent possible. The advanced data collection in the DAVID Knowledgebase not only creates a solid annotation data foundation for the various DAVID analytic tools, but also is freely available to the public in a simple pair-wise text format to promote the development of novel annotation algorithms and techniques within the scientific community. The DAVID Bioinformatics Resources are accessible at http://david.niaid.nih.gov.
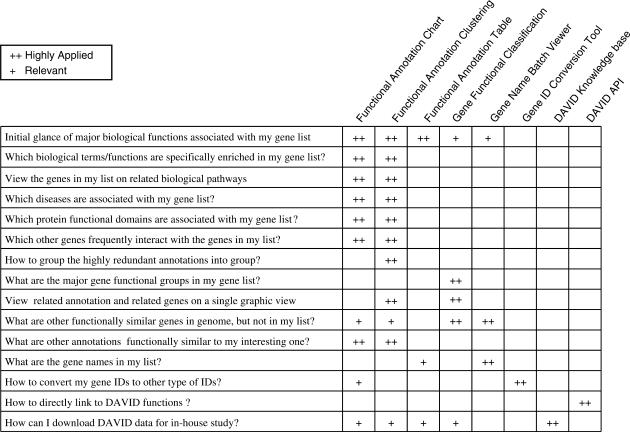
Figure 3.
A roadmap to choose appropriate DAVID functions and tools.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors are grateful to the referees and editors for their constructive comments. Thanks goes to Melaku Gedil, Ping Ren, and Jun Yang in the LIB group for biological discussions. We also thank Bill Wilton and Mike Tartakovsky for information technology and network support. This research was supported in whole by the National Institute of Allergy and Infectious Disease. This project has been funded in whole with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01- CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Funding to pay the Open Access publication charges for this article was provided by the same source as above.
Conflict of interest statement. None declared.
APPENDIX: URLS TO ACCESS MAJOR COMPONENTS IN DAVID
DAVID Home Page: http://david.niaid.nih.gov or http://david.abcc.ncifcrf.gov
DAVID Knowledgebase Download: http://david.abcc.ncifcrf.gov/knowledgbase
DAVID Functional Annotation Tool Suite: http://david.abcc.ncifcrf.gov/summary.jsp
DAVID Gene Functional Classification Tool Suite: http://david.abcc.ncifcrf.gov/gene2gene.jsp
DAVID Gene ID Conversion Tool: http://david.abcc.ncifcrf.gov/conversion.jsp
DAVID Gene Name Batch Viewer: http://david.abcc.ncifcrf.gov/list.jsp
DAVID NIAID Pathogen Browser Tool: http://david.abcc.ncifcrf.gov/GB.jsp
DAVID API Services: http://david.abcc.ncifcrf.gov/api
DAVID Forum: http://david.abcc.ncifcrf.gov/forum
REFERENCES
- 1.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 3.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 4.Berriz GF, King OD, Bryant B, Sander C, Roth FP. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 5.Bluthgen N, Brand K, Cajavec B, Swat M, Herzel H, Beule D. Biological profiling of gene groups utilizing Gene Ontology. Genome Inform. 2005;16:106–115. [PubMed] [Google Scholar]
- 6.Shah NH, Fedoroff NV. CLENCH: a program for calculating Cluster ENriCHment using the Gene Ontology. Bioinformatics. 2004;20:1196–1197. doi: 10.1093/bioinformatics/bth056. [DOI] [PubMed] [Google Scholar]
- 7.Masseroli M, Galati O, Pinciroli F. GFINDer: genetic disease and phenotype location statistical analysis and mining of dynamically annotated gene lists. Nucleic Acids Res. 2005;33:W717–W723. doi: 10.1093/nar/gki454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Hu ZZ, Wu CH. DynGO: a tool for visualizing and mining of Gene Ontology and its associations. BMC Bioinformatics. 2005;6:201. doi: 10.1186/1471-2105-6-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JS, Katari G, Sachidanandam R. GObar: a gene ontology based analysis and visualization tool for gene sets. BMC Bioinformatics. 2005;6:189. doi: 10.1186/1471-2105-6-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo-Davis CI, Hartl DL. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- 13.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 14.Zhong S, Storch KF, Lipan O, Kao MC, Weitz CJ, Wong WH. GoSurfer: A graphical interactive tool for comparative analysis of large gene sets in Gene Ontologytrade mark Space. Appl. Bioinformatics. 2004;3:261–264. doi: 10.2165/00822942-200403040-00009. [DOI] [PubMed] [Google Scholar]
- 15.Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B. GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 2004;5:R101. doi: 10.1186/gb-2004-5-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Shaul Y, Bergman H, Soreq H. Identifying subtle interrelated changes in functional gene categories using continuous measures of gene expression. Bioinformatics. 2005;21:1129–1137. doi: 10.1093/bioinformatics/bti149. [DOI] [PubMed] [Google Scholar]
- 19.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson PN, Wollstein A, Bohme U, Beattie B. Ontologizing gene-expression microarray data: characterizing clusters with Gene Ontology. Bioinformatics. 2004;20:979–981. doi: 10.1093/bioinformatics/bth040. [DOI] [PubMed] [Google Scholar]
- 21.Draghici S, Khatri P, Bhavsar P, Shah A, Krawetz SA, Tainsky MA. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 2003;31:3775–3781. doi: 10.1093/nar/gkg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatri P, Bhavsar P, Bawa G, Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatri P, Sellamuthu S, Malhotra P, Amin K, Done A, Draghici S. Recent additions and improvements to the Onto-Tools. Nucleic Acids Res. 2005;33:W762–W765. doi: 10.1093/nar/gki472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu CH, Yeh LS, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu Z, Kourtesis P, Ledley RS, et al. The protein information resource. Nucleic Acids Res. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.