Abstract
SCOPE is a novel parameter-free method for the de novo identification of potential regulatory motifs in sets of coordinately regulated genes. The SCOPE algorithm combines the output of three component algorithms, each designed to identify a particular class of motifs. Using an ensemble learning approach, SCOPE identifies the best candidate motifs from its component algorithms. In tests on experimentally determined datasets, SCOPE identified motifs with a significantly higher level of accuracy than a number of other web-based motif finders run with their default parameters. Because SCOPE has no adjustable parameters, the web server has an intuitive interface, requiring only a set of gene names or FASTA sequences and a choice of species. The most significant motifs found by SCOPE are displayed graphically on the main results page with a table containing summary statistics for each motif. Detailed motif information, including the sequence logo, PWM, consensus sequence and specific matching sites can be viewed through a single click on a motif. SCOPE's efficient, parameter-free search strategy has enabled the development of a web server that is readily accessible to the practising biologist while providing results that compare favorably with those of other motif finders. The SCOPE web server is at <http://genie.dartmouth.edu/scope>.
INTRODUCTION
The de novo identification of transcription factor binding sites is one of the oldest problems in bioinformatics with nearly a hundred algorithms published in the last 25 years (1–4). Despite the abundance of motif finders, most programs are difficult for non-expert users to readily apply to their uncharacterized datasets. Most motif-finding algorithms ask users to specify numerous parameters to describe the motifs being sought, such as length, orientation and even (in some cases) the number of expected occurrences and the expected number of genes that will contain binding sites. The existence of nuisance parameters such as these may prove frustrating for the non-expert. With many parameters to set, the user is often left to explore the parameter space and make arbitrary judgment calls on what output to trust. Many programs circumvent this issue by specifying reasonable default parameters, but studies have shown that these programs are often quite sensitive to parameters and underperform when the defaults are used (5).
In addition, the existence of nuisance parameters complicates the assessment of motif finder performance comparisons. For instance, in a recent study, thirteen motif finders were compared as run by experts (6). A number of the programs were run with different parameter settings for each regulon, and in some cases, motifs were filtered by hand both from the input sequence set and from the output sequence set. Such performance comparisons assess both the performance of the program and the expertise of the user, making it difficult for the first-time user to select a motif-finding program on a principled basis.
We recently presented a motif finder specifically developed to meet the needs of practising biologists who are interested in using motif-finding tools to identify potential transcription factor binding sites in a set of (otherwise uncharacterized) upstream regions of co-regulated genes. Our program, SCOPE (Suite for Computational identification Of Promoter Elements), requires no inputs beyond a set of unaligned sequences or gene names and a species selection. SCOPE is an ensemble learning method based on three component algorithms, each aimed at a specific category of motifs (Chakravarty et al., submitted). The component algorithms, BEAM (7), PRISM (8) and SPACER (9), are designed for the discovery of short non-degenerate motifs, short degenerate motifs and long highly degenerate (or bipartite) motifs, respectively. SCOPE combines these methods using a unified scoring metric and a ‘winner takes all’ learning rule. When we evaluated SCOPE's performance on 78 published regulons from four different species, it outperformed ten other well-known motif finders on this dataset by a large, statistically significant margin (Chakravarty et al., submitted). SCOPE was both highly sensitive and specific in its predictions, ranking in the top two out of eleven motif finders by both these criteria. Our tests also showed SCOPE to be robust to the presence of noise (extraneous genes) in test datasets, making it particularly useful for the analysis of microarray data. In semi-synthetic test regulons, where 80% of the upstream sequences were extraneous (randomly selected), SCOPE's performance degraded by only 21%.
As a cautionary note, however, our test datasets were dominated by prokaryotic and yeast regulons. Only a handful of well-characterized regulons exist for higher eukaryotes, and all motif-finding programs tested so far, including SCOPE, perform much less strongly on these organisms (6, Chakravarty et al., submitted).
This article presents the interface design and functionality of the web server for SCOPE.
WEB INTERFACE
Design philosophy
In designing the web interface for SCOPE, we sought to minimize user input while providing the maximum breadth of information as output. We adhered to the principle of revealed complexity in the interface. In keeping with this principle, only the most relevant information is provided on each output page to maximize readability and to encourage exploration by making detailed output information available with a single click of the mouse. Location-relevant help links are available on the site to facilitate ease of use.
The absence of nuisance parameters enabled us to design a clean and simple interface for input. Output pages are specifically structured to make the most commonly used information easy to view. These features result in an interface that is at once informative and simple to navigate. In addition, users can request a copy of the output through email. The emailed results are easy to parse for further analysis.
Input form
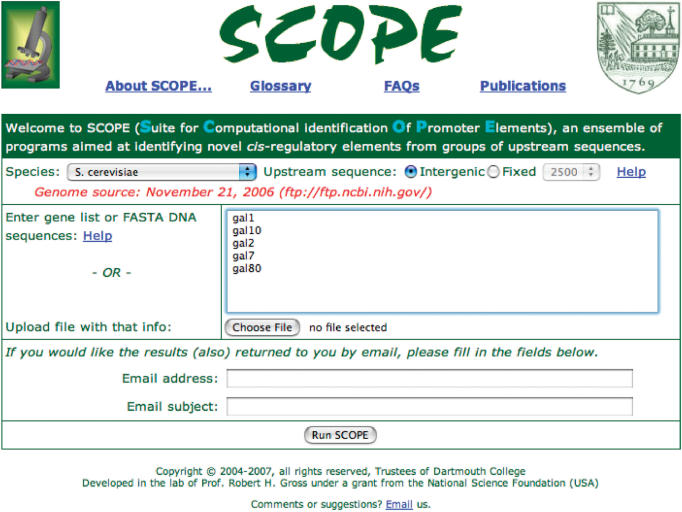
The only required input on SCOPE's input page is a list of genes (or FASTA sequences) and a species designation (Figure 1). Additionally, the user may provide an email address (and subject line) to which machine-parsible results will be sent. For gene entry, a series of FASTA sequences (or a file containing such sequences) or a list of gene names may be entered. The only input parameter that SCOPE cannot automatically optimize is the choice of input sequence length. The user may select a particular fixed length of upstream sequence to be analyzed or may select just the intergenic sequences (up to the previous gene) to be analyzed. This upstream length is also used to specify the background used in calculations of significance for both gene lists and FASTA analyses.
Figure 1.
SCOPE home page. The drop-down menu for “Species” has been used to select S. cerevisiae and the user has chosen to examine the 800 bp upstream of the transcription start site for the set of genes typed into the gene list box.
SCOPE Output
A typical run of SCOPE takes on the order of 1–5 min. Runtime is dependent primarily on the size of the genome used for the background and is rate limited by the SPACER algorithm, the slowest of the three component algorithms. SPACER's slow runtime stems from its search space, which often involves finding the exact genomic positions of a large number of short motifs. (A detailed discussion of SPACER's runtime complexity is provided in reference 9.)
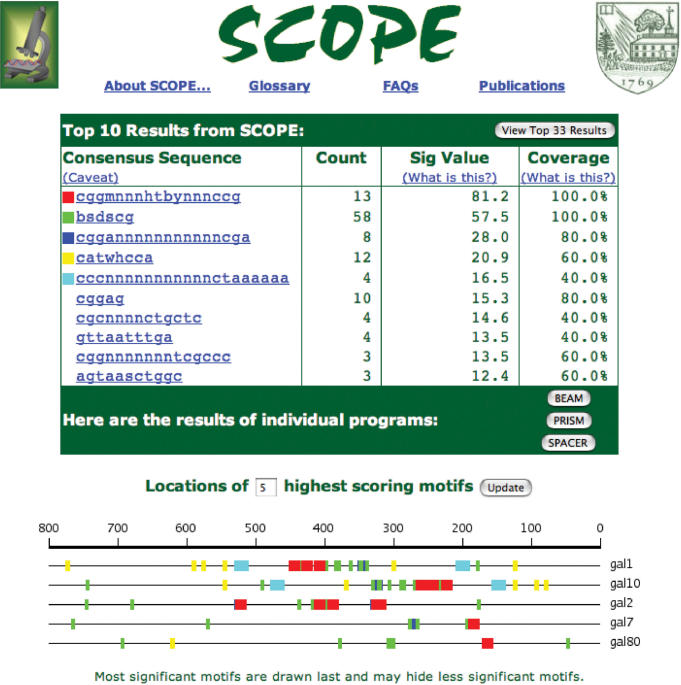
Results from SCOPE are displayed in a compact, motif-centric way (Figure 2). Initially, only the top ten motifs from SCOPE are displayed. Each motif is represented as a consensus sequence, and the sequence provides a link to more detailed information about the motif. The number of occurrences of the motif in the set of genes is also displayed along with the Sig score (a measure of the statistical significance of the motif) and the coverage (percentage of genes containing the motif). The upstream locations of the top five motifs from SCOPE are plotted in a color-coded motif map at the bottom of the page. The user can change the number of motifs drawn in the map using the available text field.
Figure 2.
Top-level results of a typical run of SCOPE. The results are shown for the gal4 regulon as entered in Figure 1. Note the motif map, which indicates that the top scoring motif (in this case, the true binding motif) is clustered in the −175 to −525 region (red).
The default view shows the combined results from each of SCOPE's component algorithms, but the individual results are available via the buttons in the bottom right corner. The individual results are provided primarily to satisfy the user's curiosity, as we have demonstrated elsewhere that SCOPE substantially outperforms each of its component algorithms (Chakravarty et al., submitted).
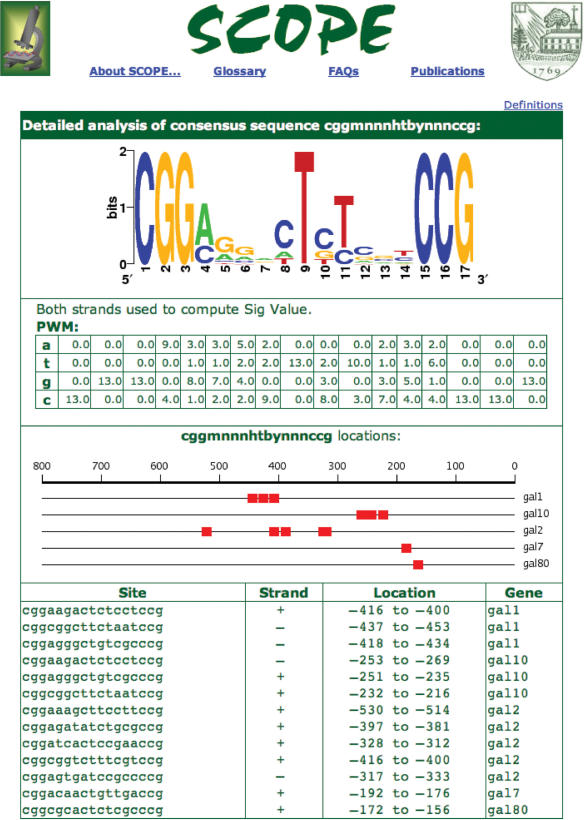
Clicking on the consensus representation of a motif on the main output page takes the user to a more detailed view of the motif (Figure 3). The additional details include the consensus sequence, the position weight matrix constructed from all instances of the motif in the regulon, a sequence logo providing a graphical view of the motif (10) and the actual instances and locations of the motif in each gene. The strand containing the motif is also displayed. For each motif, SCOPE computes the significance once considering both strands, and once considering only the top strand and the higher scoring result is displayed.
Figure 3.
Motif-level output from SCOPE. This is the page that results when the first motif in Figure 2 is clicked. The sequence logo displays additional information from what is displayed in the consensus sequence. The PWM provides the details of occurrence of each nucleotide for each position of the motif. The table at the bottom lists all of the occurrences of this motif in the upstream sequences of the set of genes submitted. This bipartite motif was identified by the SPACER algorithm, which has also helped specify some preferences for nucleotides in the internal “spacer” sequence.
Implementation
SCOPE is implemented in Java 1.4. The interface is assembled in HTML, JSP and PHP. The SCOPE server is an Apple Macintosh Workgroup Cluster for Bioinformatics.
CONCLUSIONS
The parameter-free nature of the SCOPE algorithm enables consistent predictions to be made every time by both first-time and experienced users of the program. We tested SCOPE against thirteen motif finders on a large, experimentally determined dataset consisting of 78 regulons with previously published binding sites from four organisms (Saccharomyces cerevisiae, Bacillus subtilis, Escherichia coli and Drosophila melanogaster). SCOPE's predictions on this dataset were found to be substantially more sensitive and specific than those obtained using the default configuration of any other motif finder we tested, including the three individual component algorithms (Chakravarty et al., submitted).
As with all other motif finders, one consideration in interpreting SCOPE's output is the interpretation of motif significance (Sig score for SCOPE). Although motifs with higher Sig scores represent more confident predictions by the algorithm, numerous studies have indicated the weak correlation between various definitions of statistical over-representation and biological relevance (5,6,11, Chakravarty et al., submitted). Nevertheless, we have found that SCOPE consistantly finds biologically relevant motifs among its top three predictions.
In conclusion, SCOPE is a powerful motif finder designed, through its simplicity, to be of particular use to biologists interested in cis-regulatory element prediction. This article describes an intuitive and compact web interface for SCOPE, which provides clear and concise output, based on the principle of revealed complexity.
Acknowledgements
This research was supported by a grant to RHG from the National Science Foundation, DBI-0445967. We wish to thank Nate Barney for implementation of the HTML and PHP code and for maintenance of the site. We would also like to acknowledge Radhika Khetani and Viktor Martyanov for a critical reading of the manuscript. This research reported and Open Access publications charges for this article were supported by a grant to RHG from the National Science Foundation, DBI-0445967.
Conflict of interest statement. None declared.
References
- 1.MacIsaac KD, Fraenkel E. Practical strategies for discovering regulatory DNA sequence motifs. PLoS Comput. Biol. 2006;4:e36. doi: 10.1371/journal.pcbi.0020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 3.Sandve GK, Drablos F. A survey of motif discovery methods in an integrated framework. Biol. Direct. 2006;1:11. doi: 10.1186/1745-6150-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GuhaThakurta D. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006;34:3585–3598. doi: 10.1093/nar/gkl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J, Li B, Kihara D. Limitations and potentials of current motif discovery algorithms. Nucleic Acids Res. 2005;33:4899–4913. doi: 10.1093/nar/gki791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 7.Carlson JM, Chakravarty A, Gross RH. BEAM: a beam search algorithm for the identification of cis-regulatory elements in groups of genes. J. Comput. Biol. 2006;13:686–701. doi: 10.1089/cmb.2006.13.686. [DOI] [PubMed] [Google Scholar]
- 8.Carlson JM, Chakravarty A, Khetani RS, Gross RH. Bounded search for de novo identification of degenerate cis-regulatory elements. BMC Bioinformatics. 2006;7:254. doi: 10.1186/1471-2105-7-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarty A, Carlson JM, Khetani RS, DeZiel CE, Gross RH. SPACER: identification of cis-regulatory elements with non-contiguous critical residues. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm041. in press. [DOI] [PubMed] [Google Scholar]
- 10.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein–DNA binding sites with applications to chromatin immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]