Abstract
We present the second version of Melina, a web-based tool for promoter analysis. Melina II shows potential DNA motifs in promoter regions with a combination of several available programs, Consensus, MEME, Gibbs sampler, MDscan and Weeder, as well as several parameter settings. It allows running a maximum of four programs simultaneously, and comparing their results with graphical representations. In addition, users can build a weight matrix from a predicted motif and apply it to upstream sequences of several typical genomes (human, mouse, S. cerevisiae, E. coli, B. subtilis or A. thaliana) or to public motif databases (JASPAR or DBTBS) in order to find similar motifs. Melina II is a client/server system developed by using Adobe (Macromedia) Flash and is accessible over the web at http://melina.hgc.jp.
INTRODUCTION
Transcription factor binding sites (TFBSs) play important roles in the regulation of gene expression. Extraction of a common TFBS from a set of DNA sequences is a practically important problem. Although a number of algorithms have been released so far to overcome this problem, none of them seem to be perfect (1–4). Thus, to avoid missing important motifs relying on only one algorithm or to check the effect of changing parameter values, it is useful to compare the prediction results obtained from different algorithms/parameter values. To support this function, we previously released a web tool named Melina (5). Recently, it was updated to its second version, Melina II. In Melina II, some of the integrated algorithms are replaced with more modern ones and the graphical representation is extensively improved. Melina II enables users to compare the results of promoter analysis more efficiently and easily.
OVERVIEW
Melina II allows running at most four out of five external algorithms [Consensus (6), MEME (7), Gibbs sampler (8), MDscan (9) and Weeder (10)] with users’ specified parameter values to avoid missing important motifs. MDscan and Weeder are newly added in this release. MDscan is a hybrid of two motif search strategies, word enumeration and position-specific weight matrix. Weeder adopts an enumerative pattern discovery algorithm carrying out an almost exhaustive search. The integration of algorithms based on different principles should help detecting subtle motifs and reducing the number of false positives. It may also be helpful to narrow down motif candidates or to detect alternative motifs by the combination of different algorithms and/or parameter values. Results of these algorithms are comparatively displayed with intuitive graphics (Figure 1).
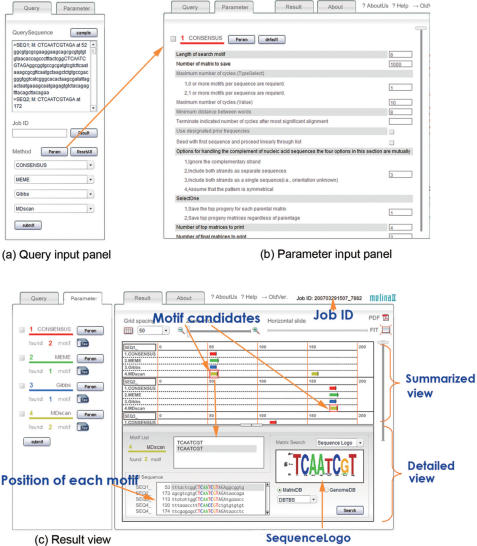
Figure 1.
Basic usage of Melina II.
As shown in Figure 1, three simple steps are sufficient to use Melina II:
Step 1: Input query sequences (Figure 1a)
In the Query input panel, multiple input sequences are fed in the FASTA format.
Step 2: Select predictive algorithms and their parameters (Figure 1a and b)
Although defaults are provided, users can choose the prediction algorithms and their specific parameter values at this step. Default parameters are sometimes chosen originally to make the search conditions as similar as possible to each other. They are: (1) the motif length is around 10 bases (‘6–10’ for MEME and Weeder; otherwise, ‘10’); (2) both strands are searched and (3) multiple occurrences are allowed for each sequence. Selecting the same algorithm with different parameter values at the same time is allowed.
Step 3: Submit a query and get results (Figure 1c)
After submitting a query, a job ID is displayed on the screen while the job is running. Users can later access the results by using this job ID.
After Melina II finishes the motif detection, the results of each prediction are integrated and displayed graphically (Figure 1c). Detected motif candidates are illustrated with colored arrows in the summarized view (upper-right corner of the result view). If users click a motif candidate in the summarized view, more information is shown in the detailed view (lower-right corner) and the predicted motif is illustrated by Sequence Logo (11) [the script for its drawing was taken from WebLogo (12)] or a weight matrix. This integrated result helps finding motif candidates and figuring out the outline of cis-regulatory modules. With the ‘PDF’ button, the output can be saved as a pdf file, which is useful either for users’ further manipulation and inclusion in publication or for getting the entire view by adjusting the scale. The ‘FIT’ button is used for conveniently getting the entire view along its horizontal axis and for hiding the detailed information at its lower half.
Furthermore, users can build a weight matrix from a predicted motif and apply it to upstream sequences of several typical genomes (human, mouse, A. thaliana, S. cerevisiae, E. coli or B. subtilis) or to public motif databases [JASPAR (13) or DBTBS (14)] in order to find similar motifs. For the former search, we used the HMMER package by Sean Eddy (http://hmmer.janelia.org/). More details are available from the help document.
EXAMPLES AND DISCUSSION
To illustrate how Melina II works, we give two examples. The first is a set of artificial DNA sequences containing several known motifs. The second consists of upstream sequences of functionally related genes.
Example 1: Embedded motifs in artificial sequences
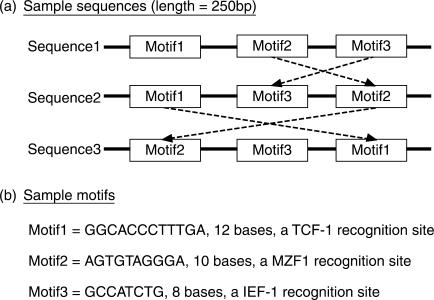
In this example, the dataset consists of three 250-bp long DNA sequences (Figure 2a). Each DNA sequence was randomly generated by the Random Sequence Generator, which is a function of Melina II. Three known consensus motifs were inserted into each sequence (Figure 2b). Motifs were set in random order to check the influence of their location. In general, it is difficult for multiple alignment programs to detect all motifs from this kind of dataset.
Figure 2.
Embedded motifs in artificial sequences.
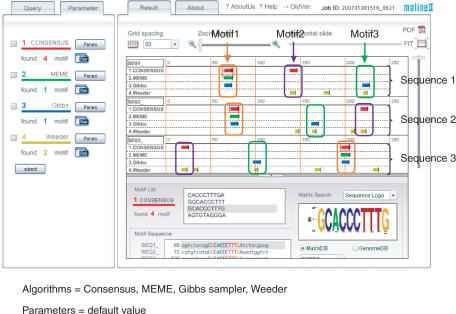
In this case, we used four algorithms, Consensus, MEME, Gibbs sampler and Weeder, with their original default parameters. This result shows that there is no predictive algorithm which can correctly detect all motifs. However, we can recover all the inserted motifs if we take motifs detected by at least two algorithms, as illustrated in Figure 3.
Figure 3.
Result view of example 1.
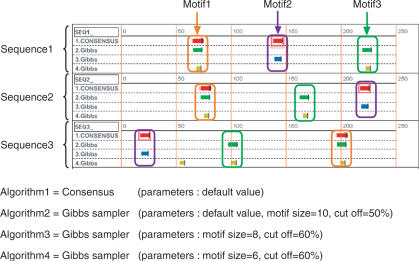
For the same dataset, we show another result in Figure 4. In this case, we used Consensus with default parameters and Gibbs sampler with three different sets of parameter values. This result clearly shows that values of parameters such as motif size and cut-off value can significantly influence motif detection. Because Melina II enables fine specification of parameters, expert users can analyze datasets multilaterally.
Figure 4.
Another result of example 1 using different parameter values.
Example 2: Upstream sequences of functionally related genes
We present here an example of real promoters containing a common motif. This dataset consists of 300 bp upstream sequences from the translational start sites of five Bacillus subtilis genes, known to be regulated by a well-known global regulator, CcpA. As shown in Figure 5, a common motif is identified and, through the search against DBTBS, it is confirmed that the motif found corresponds to the CcpA motif. (Figure 5b and c).
Figure 5.
Real promoters and motif database search.
FUTURE PROSPECTS
One future direction is to endow Melina a function to ‘guide’ favorable parameter values to improve the detection accuracy. It is not an easy task because optimal parameter values for each algorithm could depend on, say, the length and the number of input sequences as well as the nature of the pattern to be sought. Nevertheless, it seems to be possible more or less to categorize typical cases with suggested optimal parameter values for each (15).
Implementation
Melina II was developed as a web-based tool by using Adobe (Macromedia) Flash. You may need to install the Flash Plug-in beforehand.
ACKNOWLEDGEMENTS
We would like to thank all groups and authors including Gary Stormo, Bill Thompson, Charles E. Lawrence, Timothy L. Bailey, Douglas L. Brutlag, Xiaole S. Liu, Giulio Pavesi, Graziano Pesole, Boris Lenhard, Sean Eddy, Thomas D. Schneider and Steven E. Brenner that made the following algorithms freely available: Consensus, Gibbs sampler, MEME, MDscan, Weeder, RefSeq, JASPAR, HMMER and Sequence Logo/WebLogo. We thank Nicolas Sierro also for critically reading the manuscript. This work was partly supported by Grant-in-Aid for Scientific Research on Priority Areas ‘Comprehensive Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding to pay the Open Access publication charges for this article was provided by a budget from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.GuhaThakurta D. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006;34:3585–3598. doi: 10.1093/nar/gkl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tompa M, Li N, Bailey TL, Church GM, Moor BD, Eskin E, Favorov AV, Frith MC, Fu Y, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 3.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 4.Kel A, Kel-Margoulis O, Borlack J, Tchekmenev D, Wingender E. Databases and tools for in silico analysis of regulation of gene expression. In: Borlak J, editor. Handbook of Toxicogenomics. VCH Weinheim; 2005. pp. 253–290. [Google Scholar]
- 5.Poluliakh N, Takagi T, Nakai K. Melina: motif extraction from promoter regions of potentially co-regulated genes. Bioinformatics. 2003;19:423–424. doi: 10.1093/bioinformatics/btf872. [DOI] [PubMed] [Google Scholar]
- 6.Stormo GD, Hartzell GW. Identifying protein-binding sites from unaligned DNA fragments. Proc. Natl Acad. Sci. USA. 1989;86:1183–1187. doi: 10.1073/pnas.86.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of 2nd International Conference on Intelligent Systems Molecular Biology. 1994:28–36. [PubMed] [Google Scholar]
- 8.Lawrence CE, Altschul SF, Boguski MS, Neuwald AF, Liu JS, Wootton JC. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 9.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation. Nat. Biotechnol. 2002:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 10.Pavesi G, Mauri G, Pesole G. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics. 2001;32:S207–S214. doi: 10.1093/bioinformatics/17.suppl_1.s207. [DOI] [PubMed] [Google Scholar]
- 11.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makita Y, Nakao M, Ogasawara N, Nakai K. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 2004;32:D75–D77. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poluliakh N, Konno M, Horton P, Nakai K. Lecture Notes in Computer Science. Vol. 3318. Springer; 2005. Parameter landscape analysis for common motif discovery programs; pp. 79–87. [Google Scholar]