Abstract
Protein motions play important roles in numerous biological processes such as enzyme catalysis, muscle contractions, antigen–antibody interactions, gene regulation and virus assembly. Knowledge of protein flexibility is also important in rational drug design, protein docking and protein engineering. However, the experimental measurement of protein motions is often difficult, requiring sophisticated experiments, complex data analysis and detailed information about the protein's tertiary structure. As a result, there is a considerable interest in developing simpler, more effective ways of quantifying protein flexibility. Recently, we described a method, called the random coil index (RCI), which is able to quantitatively estimate backbone root mean square fluctuations (RMSFs) of structural ensembles and order parameters using only chemical shifts. The RCI method is very fast (<5 s) and exceedingly robust. It also offers an excellent alternative to traditional methods of measuring protein flexibility. We have recently extended the RCI concept and implemented it as a web server. This server allows facile, accurate and fully automated predictions of MD RMSF values, NMR RMSF values and model-free order parameters (S2) directly from chemical shift assignments. It also performs automatic chemical shift re-referencing to ensure consistency and reproducibility. On average, the correlation between RCI predictions and experimentally obtained motional amplitudes is within the range from 0.77 to 0.82. The server is available at http://wishart.biology.ualberta.ca/rci.
INTRODUCTION
NMR spectroscopy occupies a unique place among experimental methods for investigating protein dynamics. This is because it can provide site-specific information about protein motions over a large range of time scales. Over the past decade, 15N NMR relaxation experiments employing model-free analysis (1,2) have become the de facto standard used to characterize protein motions on a picosecond to nanosecond time scale. However, as with any scientific method, this approach has certain limitations (3,4). Perhaps the most obvious difficulty lies in the fact that 15N relaxation measurements are inherently time-consuming and tedious, often requiring many hours of data collection, processing and spectral analysis. A second problem is that relaxation measurements are often seriously compromised by peak overlap, poor signal intensity or peak broadening. A third problem is that the precision and accuracy of relaxation-derived measurements tends to deteriorate rapidly as the frequency of internal nanosecond motions approach that of the protein's overall tumbling rate (5,6). This is because 15N relaxation rates become insensitive to internal fluctuations that are much slower than overall tumbling. Since calculations of relaxation rates are based on measuring peak intensities, their accuracy can be severely compromised by low signal-to-noise ratios. This is especially true when dealing with larger (>150 residues) proteins or proteins undergoing μs–ms conformational exchange. Another complication to the model-free formalism lies in the fact that its proper application often requires information about anisotropy of protein overall diffusion and, as a result, the method cannot be used when the 3D structure is not known or when it is largely disordered. These limitations with traditional relaxation measurements prompted us to develop a new, chemical-shift-based technique to characterize protein mobility from NMR data. Specifically, we wanted to develop an easy-to-use, robust approach that would not be affected by protein tumbling rates, uncertainties in peak intensities or lack of knowledge about the protein's 3D structure. This method is called the random coil index or RCI (7).
As described in our previous publications (7,19), the RCI method exploits the fact that there is a remarkable amount of dynamic information intrinsic to NMR chemical shifts. The connection between chemical shifts, especially random coil chemical shifts, and protein flexibility has been known for quite some time (8–11). Random coil chemical shifts can be defined as shifts that result from a fast exchange among energy-weighted populations of all theoretically possible conformations of an unfolded polypeptide chain (12,13). The difference between an observed chemical shift for a given amino acid in a given protein, and its corresponding random coil value is called the secondary chemical shift. Secondary chemical shifts have been used for many years to qualitatively estimate the level of protein structural disorder (14–18). However, until recently, no quantitative relationships between secondary chemical shifts and protein dynamic parameters had been derived. The RCI is able to combine the chemical shift data from six different nuclei (13Cα, 13Cβ, 13CO, 15N, 1HN and 1Hα—or any combinations thereof) into a single parameter that closely correlates with amplitudes of backbone protein motions such as order parameters (S2) and root mean square fluctuations (RMSFs) of structural ensembles.
Previous descriptions of the RCI method focused on explaining the algorithm, rationalizing its utility and assessing its accuracy. As a result, only a modest effort went into making the RCI-based software user-friendly and flexible. In an attempt to make the RCI approach more accessible to the NMR community and much more robust, we decided to create an RCI web server and to optimize the RCI protocol for NMR assignments with different degrees of completeness and mis-referencing. The RCI server is a unique server, designed to support rapid (5–10 s), residue-specific determination of protein flexibility and protein mobility using only chemical shift assignment data as input. It accepts almost any combination of NMR chemical shift assignments as its input and it outputs the expected values of the RMFs of MD and NMR ensembles as well as model-free order parameters (1,2). To improve the accuracy and reproducibility of the calculations, the RCI server also supports automated chemical shift re-referencing. Assessments of the RCI server performance show an agreement between the values it calculates and experimentally obtained amplitudes of motions range from R = 0.77−0.82 depending on the type of experimental method. The RCI web server can be accessed at: http://wishart.biology.ualberta.ca/rci.
SERVER AND PROGRAM DESCRIPTION
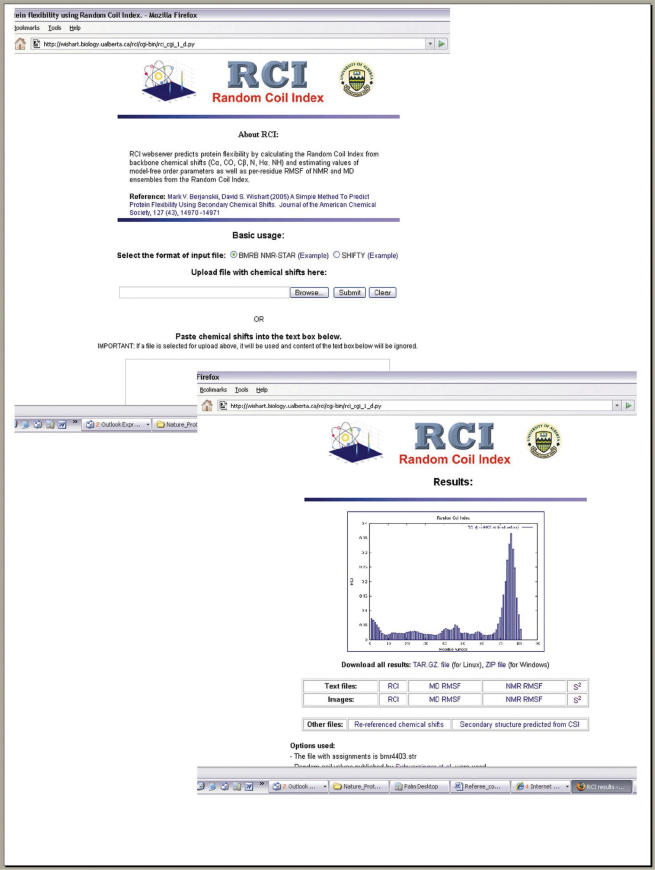
The RCI server is composed of two parts, a front-end web-interface (written in Python and HTML) and a back-end consisting of several programs including RCI (7), CSI (11), REFCOR [based on (19)], as well as several parsing and conversion utilities for handling different input files. The CSI program is written in ANSI standard C, while RCI, REFCOR, the input parsing and the conversion utilities are written in Python. The source code for the basic algorithm is available from the authors upon request. The RCI server accepts protein chemical shift assignments in standard BMRB NMR-STAR (20) and SHIFTY (21) formats as input. Users can either upload an input file into the web server (via a browse button) or paste the data in a standard text box (Figure 1). Users are also offered several options to adjust program operations to suit their specific needs. Additional details about these options and what they can offer are described later.
Figure 1.
Sample screenshots of the RCI web server's input and output pages.
A flow chart describing the basic RCI server operations is shown in Figure 2. It is also described in more detail here. To begin: (1) Experimental chemical shifts are first uploaded by the user. (2) The input chemical shifts are then re-referenced (if necessary) by REFCOR. (3) The protein sequence is extracted from the NMR assignments and used to (4) determine the appropriate random coil chemical shifts (22) and (5) determine the neighboring residue correction factors for the i ± 1 and i ± 2 residues (23). The correction factors are applied to the random coil chemical shifts to obtain reference chemical shifts. (6) Reference chemical shifts are then subtracted from the corrected experimental chemical shifts to obtain the secondary chemical shifts for the 13Cα, 13Cβ, 13CO, 15N, 1HN and 1Hα nuclei. (7) Optionally, gaps in the chemical shift assignments, if any, are filled in by averaging the chemical shifts of neighboring residues. (8) Secondary chemical shifts are smoothed by three-point moving averaging. (9) Secondary chemical shifts of are scaled to account for differences in their resonance frequencies, and, if below a certain ‘floor limit’ (currently 0.5 p.p.m.), replaced with this floor value. (10) Initial RCI values are calculated using the following expression.
 |
1 |
where |ΔδCα|, |ΔδCO|, |ΔδCβ|, |ΔδN|, |ΔδNH| and |ΔδHα| are the absolute values of the secondary chemical shifts (in p.p.m.) of Cα, CO, Cβ, N, NH and Hα, respectively. A, B, C, D, E and F are weighting coefficients (Table 1 in the RCI Online Help). Left angle and right angle brackets (<·>) indicate that the average is being calculated. (11) End-effect corrections are optionally applied (see later). (12) If the RCI values are above a certain ‘ceiling limit’ (currently 0.6), they are replaced with this ceiling value. (13) The final RCI values are obtained after a second smoothing by three-point averaging. (14) Finally, in the last step the expected values of model-free order parameters (S2), RMFs of MD and NMR ensembles are calculated using the following empirical expressions.
| 2 |
| 3 |
| 4 |
A more detailed description of the aforementioned steps has been published elsewhere (19).
Figure 2.
A flow chart describing the RCI protocol.
By default, the RCI web server uses random coil reference chemical shifts and neighboring residue correction values originally published by Schwarzinger and co-authors (22,23). This is the only set of random coil values and neighboring residue correction for residues i ± 1 and i ± 2 that were obtained under similar experimental conditions and, therefore, are expected to be quite consistent with each other. However, users can also select different sets of random coil chemical shifts, including those values published by Wang and Jardetzky (24), Wishart et al. (25) and Lukin et al. (26) using radio-buttons labeled ‘Wang’, ‘Wishart’ and ‘Lukin’, respectively. Users are also offered an option to select i ± 1 neighboring residue correction values published by Wang and Jardetzky (24) instead of the default values. While these options do affect RCI-based flexibility profiles, the differences in mean correlation coefficients for different combinations of random coil values and neighboring residue corrections (Table 2 of the RCI Online Help) are relatively small and may be considered statistically insignificant.
By default, the RCI web server does not produce predictions for unassigned residues. However, if unassigned residues are located in the middle of a well-defined secondary structure region (e.g. α-helix, β-sheet), it is not unreasonable to expect that their flexibility will be similar to that of neighboring assigned resides. In these cases, the RCI web server allows users to predict the flexibility of unassigned residues based dynamic properties of adjacent regions. This option is generally not recommended when more than two consecutive residues are unassigned.
The RCI web server also has a somewhat similar option to fill small gaps in the per-residue distributions of secondary chemical shifts. Such gaps happen more often than a lack of assignments for all the six nuclei and should be dealt with separately. The use of incomplete assignments generally reduces the quality of RCI-based flexibility predictions (see Table 1 of the RCI Online Help). By default, the RCI web server fills small gaps in per-residue distributions of secondary chemical shifts by averaging secondary chemical shifts of residues i + 1 and i − 1, and, if not available, residues i + 2 and i − 2. Users are offered an option to turn this feature off.
RCI values can also be affected by so-called ‘end effects’. These are poorly understood phenomena that alter the chemical shifts of the N- and C-termini of proteins in unpredictable ways. The RCI web server offers an option to apply an end-effect correction to the RCI values. This end-effect correction is turned on by default. A detailed protocol describing how the end-effect correction is handled was published elsewhere (27). Users should be aware that the improvement of RCI predictions due to the end-effect correction is not related to the dynamic averaging of chemical shifts. Rather, it originates from the indirect correlation between the severity of the end effects and protein flexibility due to their shared dependence on the proximity to a terminal residue. Therefore, if the correction is applied, the RCI profile at the terminal regions should be interpreted as a commonly observed trend of protein flexibility at termini of proteins. The RCI server offers users the option to disable the end effect correction or to exclude the first and the last three residues from predictions.
A particularly useful and important feature of the RCI web server is its capacity to correct mis-referenced chemical shifts. This automated reference correction is done using a local implementation of the REFCOR program. About 20% of newly deposited assignments in the BMRB database were found mis-referenced by a recent survey (28). Given the relatively high level of mis-referencing in biomolecular NMR and given the fact that mis-referenced shifts can substantially reduce the performance of chemical-shift-based methods such as RCI (7), we believed that implementing this reference correction protocol was absolutely vital to maintaining the server's performance.
The REFCOR reference correction protocol is based on the secondary structure predictions from the chemical shift index (CSI) (29) and was published elsewhere (27). Briefly, REFCOR uses CSI-based predictions of secondary structure along with typical chemical shift values observed in different secondary structures are used to calculate the necessary reference correction for each nucleus. Since CSI predictions are also affected by shift mis-referencing, the protocol is repeated several times using newly generated re-referenced shifts until the CSI predictions become stable. The reference correction option is always turned on by default although it can be switched off, if necessary.
An average RCI run takes between 5 CPU s (without chemical shift reference correction) and ∼20 s (with reference correction). An example of the web server output is shown in Figure 1. As might be expected, the RCI web server displays the name of the input file, the options selected and a plot of the per-residue RCI distribution. Users may download per-residue distributions of the RCI, predicted model-free order parameters, predicted RMSFs of MD and NMR ensembles as both text files and graphical images. These files may also be obtained individually or as a single linked file containing all the results. Re-referenced chemical shifts and CSI-based predictions of secondary structure are also available for download or direct viewing on the website. In addition to its extensive data output, the RCI web server also offers a comprehensive list of help pages to assist users in preparing their input files, in understanding the RCI method and in interpreting the web server output. This information is provided to make the RCI protocol as transparent as possible and to facilitate protocol troubleshooting if required.
PERFORMANCE AND VALIDATION
The RCI web server was optimized and evaluated on a set of 18 proteins ranging in length from 56 to 283 residues (1585 residues in total), for which complete or nearly complete 1H, 13C and 15N chemical shifts were known (Table 3 of the RCI Online Help). A subset of 14 proteins was used to generate molecular dynamic (MD) ensembles and optimize weighting coefficients in the RCI expression [Equation (1)]. This was done by using a simple grid search to maximize RCI correlation with RMSF values determined by molecular dynamics. The weighting coefficients were obtained for all 63 possible assignment scenarios (Table 1 of the RCI Online Help). To evaluate the performance of the algorithm for the training set, we used a leave-one-out procedure by removing the query protein from RCI training set prior to running the program. The average correlation coefficients for the full training set and for all leave-one-out runs were identical (R = 0.82). To ensure that the RCI algorithm had not been over-trained, four additional proteins, which were not previously included in the training set, were tested. The average correlation coefficient between the RCI determined MD RMSF and the calculated MD RMSF for these proteins was identical (R = 0.82) to that of the training set (see Table 3 of the RCI Online Help for information about the tested proteins).
To validate the relationship between RCI predictions and the amplitudes of protein motions, the correlation of RCI with model-free order parameters [observed and predicted from the structure (30)] and per-residue RMSF values of NMR ensembles were calculated. The average correlation coefficients for the eighteen sets of order parameters and sixteen sets of NMR RMSFs were 0.77 and 0.81, respectively. The performance of the conversion expressions shown in Equations (2–4) were assessed by calculating the average error between RCI-predicted motional amplitudes and the corresponding experimentally and theoretically obtained parameters. The average errors for the order parameters (18 proteins), the NMR RMSFs (16 proteins) and the MD RMSFs (18 proteins) were 0.05, 0.44 Å and 0.50 Å, respectively. Figure 1 in the RCI Online Help shows examples of the good correlations obtained between RCI and other measures of protein motional amplitudes (e.g. NMR RMSF, MD RMSF, S2) for such pharmaceutically important proteins as interleukin-4 and the HIV-1 Gag protein. Additional information about optimizing and testing the RCI method (i.e. details of MD simulations, PDB IDs and BMRB codes of proteins, etc.) can be found in the original papers on the RCI method (7) as well as on the RCI Online Help pages.
CONCLUSION
In summary, we have described a web server that is capable of rapidly and accurately predicting protein flexibility using only chemical shift assignments as the input. Comparisons suggest that these predictions correlate well with other measures of protein mobility, such as model-free order parameters and root-mean square fluctuations (RMSFs) of NMR and MD ensembles. The approach is generally applicable to proteins of any size for which 1H, 13C and 15N shift assignments are available. The web server is unique in its ability to extract dynamic information from NMR data without the prior knowledge of tertiary structure, without the need for favorable rates of rotational diffusion and without the need for exceptionally good spectral sensitivity. We have already used the RCI approach to monitor a number of interesting dynamic processes in proteins, such as the pH-induced conversion of the prion protein (PrPc) into its scrapie form (PrPsc) and the dynamic response of picornaviral protease active sites to inhibitor binding (manuscripts in preparation). We believe the RCI method may lead to important changes in the ways backbone protein flexibility is measured and reported by the scientific community.
ACKNOWLEDGEMENTS
Funding for this project was provided by PrioNet Canada, Alberta Prion Research Institute, the Protein Engineering Network of Centres of Excellence (PENCE), the Natural Sciences and Engineering Research Council (NSERC), the National Research Council (NINT) and Genome Alberta (a division of Genome Canada). Funding to pay the Open Access publication charges for this article was provided by Genome Alberta.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 2.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J. Am. Chem. Soc. 1990;112:4989–4991. [Google Scholar]
- 3.Korzhnev DM, Orekhov VY, Arseniev AS. Model-free approach beyond the borders of its applicability. J. Magn. Reson. 1997;127:184–191. doi: 10.1006/jmre.1997.1190. [DOI] [PubMed] [Google Scholar]
- 4.Case DA. Molecular dynamics and NMR spin relaxation in proteins. Acc. Chem. Res. 2002;35:325–331. doi: 10.1021/ar010020l. [DOI] [PubMed] [Google Scholar]
- 5.Jin DQ, Andrec M, Montelione GT, Levy RM. Propagation of experimental uncertainties using the Lipari-Szabo model-free analysis of protein dynamics. J. Biomol. NMR. 1998;12:471–492. doi: 10.1023/a:1008313319334. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Brooks CL, III, Wright PE. Model-free analysis of protein dynamics: assessment of accuracy and model selection protocols based on molecular dynamics simulation. J. Biomol. NMR. 2004;29:243–257. doi: 10.1023/B:JNMR.0000032504.70912.58. [DOI] [PubMed] [Google Scholar]
- 7.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 8.Grathwoh C, Wuthrich K. C-13 Nmr of protected tetrapeptides Tfa-Gly-Gly-L-X-L-Ala-Och3, where X Stands for 20 common amino-acids. J. Magn. Reson. 1974;13:217–225. [Google Scholar]
- 9.Bundi A, Grathwohl C, Hochmann J, Keller RM, Wagner G, Wuthrich K. Proton Nmr of protected tetrapeptides Tfa-Gly-Gly-L-X-L-Ala-Och3, where X Stands for one of 20 common amino-acids. J. Magn. Reson. 1975;18:191–198. [Google Scholar]
- 10.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 11.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Meth. Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 12.Bundi A, Wuthrich K. H-1-Nmr parameters of the common amino-acid residues measured in aqueous-solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-Oh. Biopolymers. 1979;18:285–297. [Google Scholar]
- 13.Vila JA, Ripoll DR, Baldoni HA, Scheraga HA. Unblocked statistical-coil tetrapeptides and pentapeptides in aqueous solution: a theoretical study. J. Biomol. NMR. 2002;24:245–262. doi: 10.1023/a:1021633403715. [DOI] [PubMed] [Google Scholar]
- 14.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. NMR analysis of a 900K GroEL GroES complex. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 15.Chou YT, Swain JF, Gierasch LM. Functionally significant mobile regions of Escherichia coli SecA ATPase identified by NMR. J. Biol. Chem. 2002;277:50985–50990. doi: 10.1074/jbc.M209237200. [DOI] [PubMed] [Google Scholar]
- 16.Lecroisey A, Martineau P, Hofnung M, Delepierre M. NMR studies on the flexibility of the poliovirus C3 linear epitope inserted into different sites of the maltose-binding protein. J. Biol. Chem. 1997;272:362–368. doi: 10.1074/jbc.272.1.362. [DOI] [PubMed] [Google Scholar]
- 17.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 18.Turner CF, Moore PB. The solution structure of ribosomal protein L18 from Bacillus stearothermophilus. J. Mol. Biol. 2004;335:679–684. doi: 10.1016/j.jmb.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Berjanskii M, Wishart NMR: prediction of protein flexibility. Nature Protoc. 2006;1:683–688. doi: 10.1038/nprot.2006.108. [DOI] [PubMed] [Google Scholar]
- 20.Seavey BR, Farr EA, Westler WM, Markley JL. A relational database for sequence-specific protein NMR data. J. Biomol. NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Watson MS, Boyko RF, Sykes BD. Automated 1H and 13C chemical shift prediction using the BioMagResBank. J. Biomol. NMR. 1997;10:329–336. doi: 10.1023/a:1018373822088. [DOI] [PubMed] [Google Scholar]
- 22.Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRview. J. Biomol. NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- 23.Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Jardetzky O. Investigation of the neighboring residue effects on protein chemical shifts. J. Am. Chem. Soc. 2002;124:14075–14084. doi: 10.1021/ja026811f. [DOI] [PubMed] [Google Scholar]
- 25.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 26.Lukin JA, Gove AP, Talukdar SN, Ho C. Automated probabilistic method for assigning backbone resonances of (C-13,N-15)-labeled proteins. J. Biomol. NMR. 1997;9:151–166. doi: 10.1023/a:1018602220061. [DOI] [PubMed] [Google Scholar]
- 27.Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. J. Biomol. NMR. 2003;25:173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 29.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Bruschweiler R. Contact model for the prediction of NMR N-H order parameters in globular proteins. J. Am. Chem. Soc. 2002;124:12654–12655. doi: 10.1021/ja027847a. [DOI] [PubMed] [Google Scholar]