Abstract
In silico screening methods based on the 3D structures of the ligands or of the proteins have become an essential tool to facilitate the drug discovery process. To achieve such process, the 3D structures of the small chemical compounds have to be generated. In addition, for ligand-based screening computations or hierarchical structure-based screening projects involving a rigid-body docking step, it is necessary to generate multi-conformer 3D models for each input ligand to increase the efficiency of the search. However, most academic or commercial compound collections are delivered in 1D SMILES (simplified molecular input line entry system) format or in 2D SDF (structure data file), highlighting the need for free 1D/2D to 3D structure generators. Frog is an on-line service aimed at generating 3D conformations for drug-like compounds starting from their 1D or 2D descriptions. Given the atomic constitution of the molecules and connectivity information, Frog can identify the different unambiguous isomers corresponding to each compound, and generate single or multiple low-to-medium energy 3D conformations, using an assembly process that does not presently consider ring flexibility. Tests show that Frog is able to generate bioactive conformations close to those observed in crystallographic complexes. Frog can be accessed at http://bioserv.rpbs.jussieu.fr/Frog.html.
INTRODUCTION
Drug discovery is a complex and expensive endeavor that has taken advantage of the recent years’ emergence of the ‘in silico’ biology. More specifically, virtual or in silico screening, based on the 3D structure of known ligands or of the targets is becoming a method of choice to facilitate lead compound identification, as seen in several recent studies [see for examples (1–5)]. In all situations, these in silico processes require a suitable compound collection as input. Usually, libraries have to be filtered (ADME/tox filtering) to remove compounds with unacceptable physico-chemical properties and disease-causing chemical functionalities. Then, the 3D structure of each compound has to be generated since, for the time being, academic or commercial compound collections are delivered in 1D SMILES (6) (simplified molecular input line entry system), CANSMILES (7) (canonical smiles) or in 2D SDF (8) (structure data file) formats. In addition, for drug design programs that use rigid-body docking steps or for 3D ligand-based screening experiments, one single conformation per compound is not enough and one has to generate conformational isomers. Very few free on-line tools are available to generate the 3D conformation of compounds. To overcome this limitation, sites such a Zinc (9), FAF-drugs (10) or very recently pubChem (11) take advantage of commercial software to propose the pre-calculated collections of compounds in 3D. Alternative services that provide direct 2D to 3D facilities come from demos of drug design package vendors, such as OpenEye’ Omega (http://www.eyesopen.com/products/applications/omega.html), Molsoft (http://www.molsoft.com/2dto3d.html), Corina (http://www.molecular-networks.com/software/corina/) and from academic sites such as (http://iris12.colby.edu/%7Ewww/jme/smiledg.html; http://davapc1.bioch.dundee.ac.uk/programs/prodrg/; http://bioserv.cbs.cnrs.fr/HTML_BIO/APPLET_ACD/create_molecule.html) (see www.vls3d.com for a more complete list). Such services are, however, usually limited to building one compound at a time and provide generally only one conformer (only the Omega's service is able to return an ensemble of conformations for the input compound).
It is well known that generating an accurate 3D structure for a small chemical compound is not trivial and as such many different approaches have been developed over the years, starting from manual model building up to quantum mechanical calculations. Between these two extremes and because for quantum chemistry programs it is generally necessary to start from a reasonable initial 3D structure, other methods such as rule-based methods (approaches based essentially on structural data) or data-based methods are commonly used (12–13).
Frog (a mixed rule-based data-based approach) aims at providing on-line generation of ensembles of 3D conformation for drug-like compounds (i.e. compounds that are ADME/tox compliant). It is based on Frowns (a chemoinformatics toolkit available at http://frowns.sourceforge.net/) to which several functionalities have been added to allow the generation of 3D structures starting from SMILES or SDF data input. Frog is able to (i) fully or partially disambiguate compound stereochemistry including chiral sites, and to (ii) generate single or ensembles of low to medium energy 3D conformations for each isomer.
CONCEPTS AND METHODS
Compound analysis, chirality assignment
The initial compound analysis performed by Frowns does not identify chiral centers not explicitly notified in the input data. New functionalities have therefore been added to detect chiral centers for the tetragonal carbons and nitrogens—for nitrogens, we have chosen to consider the chiral forms instead of the average plane conformation that is sometimes employed, since it can have some impact on 3D coordinate generations. Briefly, to identify chiral centers, we take advantage of the cansmiles representation generated by Frowns. The cansmiles of the groups bonded to the putative chiral center are extracted and compared. Chiral centers bonded group cansmiles are all different. Note that difficult cases involving symmetries can be managed simply by such strategy. ZE chirality for double bonds as well as axial/equatorial positions is treated in the same way.
3D assembly of an initial conformer
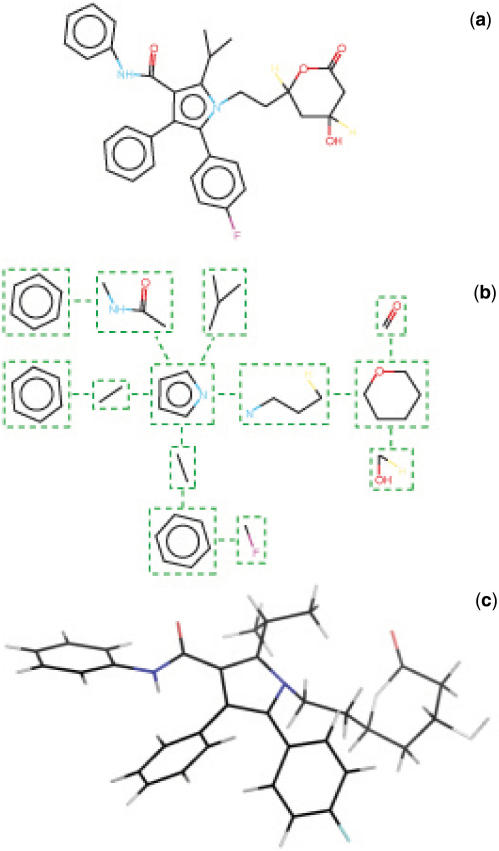
One difficult task in the generation of 3D conformation for drugs has long been identified as coming from the ring systems (12). As a result, 3D structure generators often fragment compounds into ring systems and acyclic parts and treat each subgroup (i.e. rings, linkers) differently. The conformational variability of ring systems cannot be handled simply by varying some torsional angles. To circumvent this problem with the rings, software such as Omega (OpenEye Scientific Software), usually make use of libraries of pregenerated ring structures but can also construct fragments on-the-fly, when needed. Acyclic parts can be handled in a much more standard manner, considering that bond length and angles can be set to standard values as reported in tables obtained by structural analysis of experimentally resolved structures. For these linker segments, conformational variability can be reduced to that of the flexible dihedral angles of the system. Frog follows such strategy: given a compound with 1D or 2D descriptions, the molecule is fragmented as a graph of rings and acyclic elements as illustrated in Figure 1. From this graph, the general strategy for the re-construction of the molecule is as follows: (i) ring conformations are taken from a library. This library consists of the conformations of the rings as extracted from the few 3D compound collections freely available on the internet. Presently, the library encompasses close to 10 100 distinct cycles. Although all the occurring conformations of each ring have been stored, Frog presently only considers the first one, i.e. rings have no conformational variability; (ii) the compound is assembled by building from scratch the acyclic components. This process is achieved on the basis of the canonical bond lengths and valence angles described in (14), supplemented by some values taken from the merck molecular force field (MMFF) (15–19) (for instance fluoride, hydrogen parameters are not provided in (14)). The junction between acyclic components and rings requires special treatment to guarantee correct chirality but follows the same basic principles. Frog can detect cycles for which axial and equatorial conformations may occur and generate both. Cycles are superimposed to the extremities of the acyclic parts using a rigid best fit procedure (20). Note that Frog generates hydrogen coordinates but does not perform pKa predictions, and as such the users may have to delete or add hydrogen atoms with the OpenBabel -p -d options (see http://openbabel.sourceforge.net/wiki/Main_Page).
Figure 1.
Flowchart of Frog processing. The compound (a) is decomposed as a graph involving rings and acyclic elements (b). Acyclic elements are built from scratch. Ring conformations are extracted from a library of rings. Then all the elements are assembled to produce the complete compound structure (c).
3D conformational variability
The exploration of the conformational variability of the compounds occurs once a first conformation is built. Only the dihedral angles of the system that are assigned to flexible covalent bonds are explored and here only heavy atoms are considered. Based on the atomic types, Frog will consider the canonical angle values of the rotatable bonds, and explore the combinations of these values. Since canonical values can result in high-energy conformations, each combination is supplemented by a few Monte-Carlo steps using a small angular perturbation (usually <10°). Such protocol was devised from several test simulations for which we checked acceptance rate and convergence towards low-energy structures. For difficult cases, this value can be increased up to 60° for angles having a 3-fold energy barrier, or to 180° for angles involving cis/trans conformations, such as the peptide bond. The conformations are scored using the Van der Waals energy of the system computed with an in-house implementation of the MMFF force field. Atomic type assignments within Frog have been checked and validated against the MMFF validation suite (http://www.ccl.net/cca/data/MMFF94s/). For compounds that have a large number of rotatable bonds the combinatorial is randomly truncated to remain tractable. Truncation also occurs for compounds having a large number of isomers. Truncating a combinatorial search is a difficult task. Different strategies have been considered. Currently, Frog randomly selects among the isomers (uniform law), and then randomly selects combinations of rotations distributed on all rotatable bonds. The Monte-Carlo process is applied to the complete set of rotatable bonds. No final energy minimization is performed, and as a consequence, some compounds have strained energy conformations. This is partially acceptable because small molecule co-crystallized with proteins can also have relatively high-energy conformations (4–15 kcal/mol above the estimated lowest energy state), while we are working on implementing a minimizer to optimize the geometry of these tense molecules.
PERFORMANCES
To assess the functionalities of Frog, several series of tests have been performed. The first one is related to the identification of chiral centers. To check that a correct identification is performed, we have taken all the compounds from the Asinex collection available at FAF-drugs as smiles, and we have removed any chirality information (including ZE information). Then we searched for the presence of chiral centers using Frog and checked the retrieval of the centers as specified in the collections. For over 84 812 compounds present in the collection, the described centers were identified for 84 792 compounds. Manual inspection of the 20 problematic cases revealed inconsistencies due to erroneous SMILES present in the collections.
The assessment of the correctness of 3D conformation assembly is more complex. First we have checked the quality of the geometry on all the isomers of series of compounds randomly taken from the Asinex, Specs and ChemBridge collections (over several hundred isomers). This visual inspection was satisfactory, including the axial/equatorial conformations.
Finally, a last test was carried out to assess the diversity and the relevance of conformations generated for one given isomer. Figure 2 shows examples of conformations generated using Frog for one compound selected among many others from our FAF-Drug test set, i.e. molecules for which the experimental conformation of the drug in situ is known (bioactive conformation). As can be seen in this figure, Frog was able to generate conformations for the raloxifene molecule that are close to the co-crystal structure. These predicted structures were not the lowest energy conformations generated during the run, and for the best one has an RMSd of 1.30 Å when compared with the bioactive conformation. Indeed, it is known that when a ligand binds to a protein, it is typically not in the lowest-energy conformation found for the unbound structure. Since we are interested here in generating 3D structures of small molecules to perform docking studies or 3D ligand-based screening experiments, it was important for us to in fact be able to generate conformations close to the bound ligand conformations. We have obviously evaluated Frog for many compounds including the new Astex validation set (21). This test set contains 85 high-resolution protein-ligand crystal structures, for which the ligand is fully drug like. First, we have assessed the performance of Frog for the generation of conformations close to those observed in experimentally resolved complexes. We have generated series of up to 50 conformers for the 85 compounds, imposing the experimental stereoisomery by converting the compound from the mol format of the library to smiles using the OpenBabel package (22). For 18 compounds, several stereoisomers were considered due to ambiguous chiral nitrogens. A full description of the results is accessible from the Frog Help page. On average, Frog closest conformation to the crystallographic conformation deviates by 1.07 Å RMS deviation. The average range from the lowest to the highest RMS deviations between the experimental structures and the conformers generated by Frog is 1.26 Å. These numbers are stable over several independent runs.
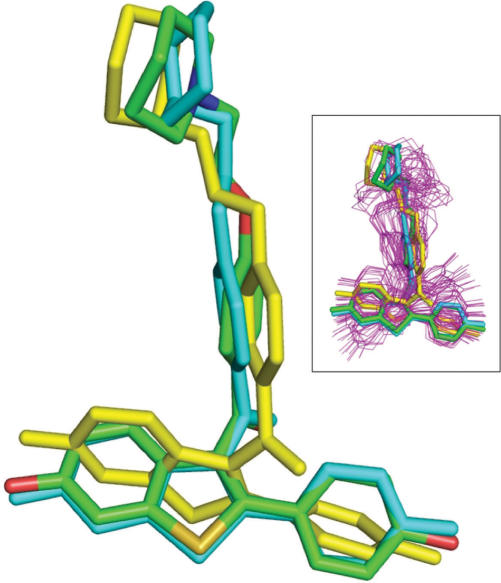
Figure 2.
Predicted versus experimental structure of raloxifene. The structure of raloxifene (PDB code 1err) (23) was predicted with FROG from a 2D SDF input file or a SMILES string. Several conformers were generated. All atom colors: experimental structure. Yellow: lowest energy structure. Cyan: conformation fitting the best the experimental structure (RMSd 1.2 Å). Inset: the 50 predicted conformations (magenta) superimposed onto the experimental structure with the experimental, lowest energy and best rmsd in stick representation.
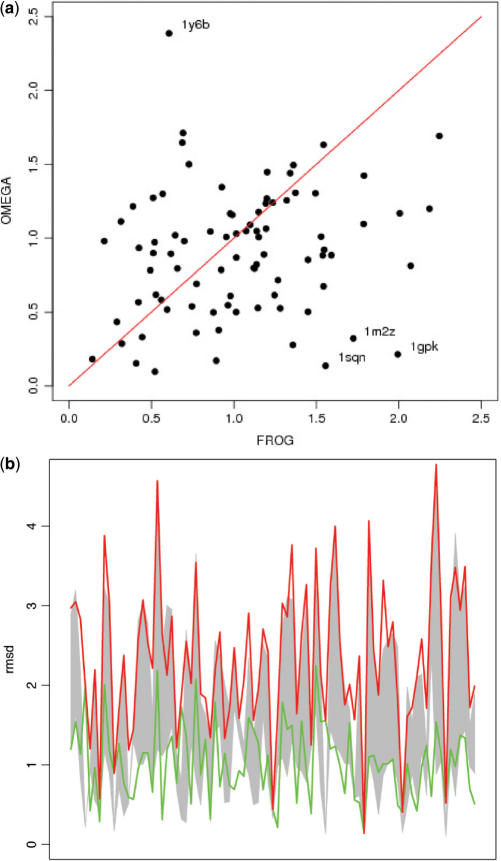
Furthermore, we also compared Frog and Omega results. With Omega, we have also generated up to 50 conformations for each 85 high-quality molecules from the Astex set and using the 3D mol files as input. As shown in Figure 3, the relative performances of Frog and Omega are close, both in terms of the conformation closest to the crystallographic conformation (Figure 3a), and of the diversity of the conformations generated (Figure 3b). Using Omega, the average deviation of the best conformation to the experimental one is of 0.90 Å, with an average range of the RMS deviation of 1.24. As shown in Figure 3a, some compounds lead to singular deviations depending on the method used. Some of these deviations can clearly be attributed to possible differences in the conformational sampling due to the Monte-Carlo method. For some other compounds such as 1gpk, 1sqn and 1m2z, we find that the conformation of the cycles from the library is mostly responsible for the large deviations. Imposing the experimental conformation of the cycle can have a dramatic impact. For instance, the best approximation for 1m2z falls from 1.7 Å down to 0.6 Å. For 1gpk and 1sqn, since the molecules contain essentially ring systems, the effect can be even more pronounced. This highlights one present limitation of Frog.
Figure 3.
(a) Deviation of the best conformation generated using Frog versus the best conformation generated by Omega, for series of up to 50 conformers. The best conformations are expressed in terms of RMS deviation to the experimental structure of the compound. The results are presented for the 85 protein-ligand crystal structures of the new Astex validation set (21). Labels identify some compounds with singular deviations (see text). (b) Diversity of the conformations generated by Frog and Omega for 85 compounds. Conformation deviations correspond to RMS deviation to the experimental structure of the compound. Omega: gray filled area; Frog: area between the red and green lines.
INPUT/OUTPUT
Frog accepts as input all compounds described using the SMILES or SDF formats. A facility is proposed to interconvert molecules to these formats using the OpenBabel package (22). Once the small molecules are in SMILES or SDF format, the possible processings are:
to generate unambiguous SMILES expression
to generate only one conformation per compound (not considering all the isomers)
to generate one conformation per isomer
to generate multiple conformations per isomer. The maximal number of multiple conformation per isomer can be selected by the user, but cannot be larger than 100. Along the same line, a maximum energy value can be specified by the users (it corresponds to the maximal energy difference to the conformation of minimal energy generated).
To keep computational effort reasonable, we presently limit the number of compounds per request to 1000. Results are on the form of a log reporting the processing of each compound, a SMILES file listing the unambiguate SMILES of the conformations generated, and a 3D file in the mol2 or SDF or PDB format.
DISCUSSION AND FUTURE DIRECTION
Frog aims at providing free on-line generation of 3D multi-conformation for series of compounds. Several features, in particular related to the disambiguation of isomers, have, to the best of our knowledge, no on-line equivalent. However, several improvements can be considered. Indeed, limitations occur from the multiconformation generator. First, the current default strategy is oriented towards exhaustive conformational space sampling. Frog attempts the systematic generation of all possible conformations of the preferred angular values of all the rotatable bonds when tractable. When this is not possible, either because there is a large number of isomers, or since the angular combinatorial to be explored becomes too large, it will perform random selection of isomers and rotatable angles. Since this process can fail and result in proposing only conformations with relatively high energy, we have implemented a Monte-Carlo procedure limited to few steps. Such process could obviously be enhanced in several ways, in particular by identifying dead end combinations of angles or isomers. This should also result in accelerated processing capabilities. Further improvement could also be expected from the force field. Conformation scoring is presently based on Van der Waals but introducing simple Coulombic electrostatics calculation could be of interest. It seems desirable to consider ring conformational variability in a next version of Frog. Also, compounds containing rings not present in the library cannot be predicted by Frog at present. This problem will be addressed in the future. Another point to consider is related to the quality of the conformations generated. While Frog will generate multiple conformations, it is possible that some of the returned conformations are close, either since several combinations of angles can produce conformations having low RMS deviations, or because of the Monte-Carlo steps. Diversity could be improved by clustering a larger number of conformations and returning only the centroids of the unrelated conformational classes generated. Nevertheless, because we focus on drug-like molecules, which limits the chemical variability of the compounds that should be treated, we have found that the initial 3D structures of numerous drug-like molecules can indeed be accurately predicted with the present version of Frog.
ACKNOWLEDGEMENTS
This RPBS project was supported by INSERM recurrent funding to U726 and U648. Funding to pay the Open Access publication charges for this article was provided by INSERM and Université Denis Diderot - Paris 7.
Conflict of interest statement. None declared.
REFERENCES
- 1.Congreve M, Murray CW, Blundell TL. Structural biology and drug discovery. Drug Discov. Today. 2005;10:895–907. doi: 10.1016/S1359-6446(05)03484-7. [DOI] [PubMed] [Google Scholar]
- 2.Hardy LW, Malikayil A. The impact of structure-guided drug design on clinical agents. Curr. Drug Discov. 2003;15:15–20. [Google Scholar]
- 3.Abagyan R, Totrov M. High-throughput docking for lead generation. Curr. Opin. Chem. Biol. 2001;5:375–382. doi: 10.1016/s1367-5931(00)00217-9. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez JC. High-throughput docking as a source of novel drug leads. Curr. Opin. Chem. Biol. 2004;8:365–370. doi: 10.1016/j.cbpa.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Schneider G, Bohm HJ. Virtual screening and fast automated docking methods. Drug Discov. Today. 2002;7:64–70. doi: 10.1016/s1359-6446(01)02091-8. [DOI] [PubMed] [Google Scholar]
- 6.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 7.Weininger D, Weininger A, Weininger JL. SMILES. 2. Algorithm for generation of Unique SMILES Notation. J. Chem. Inf. Comput. Sci. 1988;29:97–101. [Google Scholar]
- 8.Dalby A, Nourse JG, Hounshell WD, Gushurst AKI, Grier DL, Leland BA, Laufer J. Description of several chemical structure file formats used by computer programs developed at Molecular Design Limited. J. Chem. Inf. Comput. Sci. 1992;32:244–255. [Google Scholar]
- 9.Irwin JJ, Shoichet BK. ZINC – a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miteva MA, Violas S, Montes M, Gomez D, Tuffery P, Villoutreix BO. FAF-Drugs: free ADME/tox filtering of compound collections. Nucleic Acids Res. 2006;34:W738–W744. doi: 10.1093/nar/gkl065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadowski J, Gasteiger J. From atoms and bonds to three-dimensional atomic coordinates: automatic model builders. Chem. Rev. 1993;93:2567–2581. [Google Scholar]
- 13.Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL. Recent developments of the chemistry development kit (CDK) – an open-source java library for chemo- and bioinformatics. Curr. Pharm. Des. 2006;12:2111–2120. doi: 10.2174/138161206777585274. [DOI] [PubMed] [Google Scholar]
- 14.Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Cryst. 1991;A47:392–400. [Google Scholar]
- 15.Halgren T. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996;5&6:490–519. [Google Scholar]
- 16.Halgren T. Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J. Comput. Chem. 1996;5&6:520–552. [Google Scholar]
- 17.Halgren T. Merck molecular force field. III. Molecular geometries and vibrational frequencies for MMFF94. J. Comput. Chem. 1996;5&6:553–586. [Google Scholar]
- 18.Halgren T. Merck molecular force field. IV. Conformational energies and geometries for MMFF94. J. Comput. Chem. 1996;5&6:587–615. [Google Scholar]
- 19.Halgren T. Merck molecular force field. V. Extension of MMFF94 using experimental data, additional computational data, and empirical rules. J. Comput. Chem. 1996;5&6:616–641. [Google Scholar]
- 20.Zuker M, Somorjai RL. The alignment of protein structures in three dimensions. Bull. Math. Biol. 1989;51:55–78. doi: 10.1007/BF02458836. [DOI] [PubMed] [Google Scholar]
- 21.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 22.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner JK, Willighagen E. The Blue Obelisk – interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]