Abstract
PrimerZ (http://genepipe.ngc.sinica.edu.tw/primerz/) is a web application dedicated primarily to primer design for genes and human SNPs. PrimerZ accepts genes by gene name or Ensembl accession code, and SNPs by dbSNP rs or AFFY_Probe IDs. The promoter and exon sequence information of all gene transcripts fetched from the Ensembl database (http://www.ensembl.org) are processed before being passed on to Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) for individual primer design. All results returned from Primer 3 are organized and integrated in a specially designed web page for easy browsing. Besides the web page presentation, csv text file export is also provided for enhanced user convenience.
PrimerZ automates highly standard but tedious gene primer design to improve the success rate of PCR experiments. More than 2000 primers have been designed with PrimerZ at our institute since 2004 and the success rate is over 70%. The addition of several new features has made PrimerZ even more useful to the research community in facilitating primer design for promoters, exons and SNPs.
INTRODUCTION
Polymerase chain reaction (PCR) is a commonly used laboratory procedure nowadays for a variety of tasks, such as DNA cloning, sequence determination and SNP detection. Consequently, numerous primers need to be designed for DNA amplification in order to produce enough DNA.
For DNA sequencing, to design primers for the promoter and exon regions of a gene, one needs to retrieve the required sequence information for each single exon and promoter, including their corresponding flanking sequences, convert them to the correct format, switch to a primer design application like Primer 3 (1), import the sequence information and adjust parameter settings if necessary before submitting the request. While this is tolerable for primer design for a gene that has a single exon, a human, mouse or rat gene can easily have more than 10 exons, and each of the above steps may thus need to be repeated 10 times or more. Furthermore, if an exon is too long to sequence properly in one run, several primers have to be designed to map overlapping sections of the exon. The whole process requires many manual steps for repeated window opening, browsing, application switching, copying and pasting, typing and so on, which can be tedious, time-consuming and error prone. In fact, for a gene with 10 exons, the process can easily exceed 300 steps.
To improve the situation, we have developed PrimerZ to replace almost all of these manual steps so that users can easily complete a primer design task using just a few clicks for a gene or batched human SNPs. More than 2000 primers have been designed with PrimerZ at our institute since 2004 and the success rate is over 70%.
PrimerZ accepts either gene names or SNP IDs (2). There are a multitude of user-definable options available including product size, maximum exon length, excluded regions of the query sequence, GC-content and maximum allowable local alignment score. Simply by submitting a candidate gene name, a SNP ID or up to 100 batched SNP IDs, in most cases all the primer sequences should be returned in a minute or two. The generated information, including gene transcript graph, primer data from Primer3, and direct links to UCSC In-Silico PCR (3,4) for PCR product prediction, to NCBI Blast and to Ensembl source data, is integrated and displayed on a single page for convenient viewing. Additionally, all the primer data can be exported in csv format for further processing.
IMPLEMENTATION
PrimerZ takes advantage of the well-developed public-domain database Ensembl, through its API (application programming interface). When a gene query is received, PrimerZ will access the Ensembl database through ENSj API (5) to retrieve promoter and exon information.
By default but with an adjustable setting, PrimerZ retrieves one region of 1440 bp upstream from the start of 5′-UTR which it treats as the promoter region. The promoter region is thereafter divided into four 360 bp non-overlapping segments plus their respective flanking sequences. The default length of flanking sequence is 240 bp. All exons are directly flanked with 240 bp sequences, except when an exon of the gene is >360 bp, when the sequence will be split into segments of ≤360 bp for a better quality sequencing result. For example, an exon of 1000 bp will produce two 360 bp segments and one 280 bp segment.
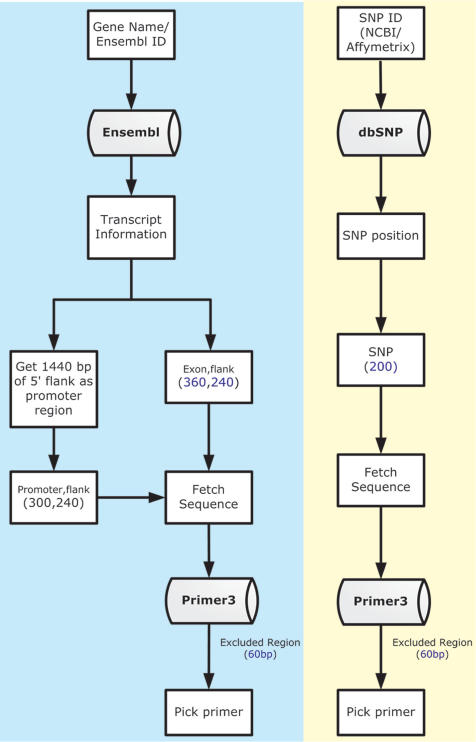
The above sequence information, plus parametric settings packaged by PrimerZ are then fed into Primer3 for primer design. All returned primer results, together with a transcript graph, are integrated into a one-page report for final output. The self-explanatory workflow of the whole PrimerZ system is shown in Figure 1.
Figure 1.
PrimerZ workflow.
The software development environment included the following software:
Java, JDK: j2sdk1.4.2_06, Server VM
Struts Framework 1.2
Red Hat Enterprise Linux Academic Edition
MySQL 4.1
Tomcat 5.0 web server
Ensembl Java API
The API provided by Ensembl is used for gene information retrieval. JAVA language is used to pipeline gene data into Primer 3 and merge the returned results.
INPUT
PrimerZ is an easy-to-use tool to design primers for genes and SNPs, using only a few simple steps to design the wanted primers. It currently provides gene primer design for all Ensembl species while SNP primer design is currently only available for human SNPs. For gene primer design, there are some essential parameters required for optimal design of primers, such as maximum exon length, exon flanking region, product size range and excluded region. Users can modify these parameters or even full range Primer 3 parameters under ‘Advance Options’ according to the particular requirements of their experiments.
Exons with length larger than the ‘maximum length’ parameter will be fragmented. The ‘excluded region’ value allows the program to bypass regions with low sequence quality or containing repetitive elements such as ALUs or LINEs for primer design. For SNP primer design, a SNP rsID or an AFFY_ProbID(6) as well as a batch file containing mixed ID types can be accepted as input, where the maximum number of SNPs per batch is limited to 100.
OUTPUT
The result page comprises three major parts: initial input data and parameters, the gene transcript diagram (only available for gene primer design) and designed primer information. The first part lists all the input data and parameters plus Ensembl Database version, for ease of reference. The second part shows each promoter fragment and exon of a transcript diagram, and links to their accompanying original Primer3 output. Finally, the third section presents tabulated primer information and incorporates executable links to NCBI blast and UCSC In-Silico PCR to check specificity and product prediction. In addition, an ‘Ensembl Link’ button offers the Ensembl Exon Report of the transcript so that a user can trace all primers to their original sequence information. A csv format text file of the results can be downloaded at this results page.
BENCHMARK
In a manual operation benchmarking test, it took a person very familiar with all processes about 1260 s and 380 discrete steps to design the primers of HADHSC (l-3-hydroxyacyl-Coenzyme A dehydrogenase, short chain), our benchmark gene with 10 exons. With automated PrimerZ, the same operation took only 16 s and three steps. This comparison clearly demonstrates the superior efficiency and ease-of-use of PrimerZ for gene primer design. A benchmark test on batched SNPs showed a similar dramatic reduction in workload from 90 discrete steps to two steps, from 840 to 49 s, and from 40 result pages to a single page.
CONCLUSION
PrimerZ has been designed to obtain reliable primers for PCR experiments and to allow standardized, automated primer design for batch operation. Users can access the UCSC In-Silico PCR directly from the result page to verify their primers to achieve higher accuracy and lower cost. PrimerZ also allows users to modify the conditions of primer design, including the maximum exon size, the flanking region of the target sequence, the exclusion region and the maximum allowed polyA and CA-repeats in the PCR products. In addition, PrimerZ will offer primer design from NCBI transcripts in the near future, which should be of great interest to those users who use NCBI data to design primers. The results from NCBI and Ensembl will be shown in the same page. Following the release of Primer3 Web Interface in November 2006, we are installing and testing a local copy of Primer3 Web to alleviate the burden on the original Primer3 website and the restriction on the number of SNPs allowed.
Primer Z is a simple-to-use program that greatly facilitates and enhances the traditionally time-consuming task of accurate primer design for PCR, and should be an excellent additional tool for the modern molecular biologist.
ACKNOWLEDGEMENTS
Special thanks to Dr Mike Lee for constructive discussions at the beginning of this work and to Dr Harry Wilson for manuscript editing. The PrimerZ project is supported by National Science Council, Taiwan, under Grant no. NSC95-3112-B-001-011(National Genotyping Center), NSC94-3112-B-001-008-Y and Academia Sinica Life Sciences Grant No. 40-19. Funding to pay the Open Access publication charges for this article was provided by National Science Council, Taiwan, under Grant no. NSC95-3112-B-001-011(National Genotyping Center).
Conflict of interest statement. None declared.
REFERENCES
- 1.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Meth. Mol. Biol. 2000;132:365. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 2.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamy P, Andersen CL, Wikman FP, Wiuf CP. Genotyping and annotation of Affymetrix SNP arrays. Nucleic Acids Res. 2006;34:e100. doi: 10.1093/nar/gkl475. [DOI] [PMC free article] [PubMed] [Google Scholar]