Abstract
The VISTA portal for comparative genomics is designed to give biomedical scientists a unified set of tools to lead them from the raw DNA sequences through the alignment and annotation to the visualization of the results. The VISTA portal also hosts the alignments of a number of genomes computed by our group, allowing users to study the regions of their interest without having to manually download the individual sequences. Here we describe various algorithmic and functional improvements implemented in the VISTA portal over the last 2 years. The VISTA Portal is accessible at http://genome.lbl.gov/vista.
INTRODUCTION
Comparing genomic sequences across related species is a fruitful source of biological insight. Functional elements such as exons tend to exhibit significant sequence similarity due to purifying selection, whereas regions that are not functional tend to be neutrally evolving and thus less conserved. The first step in comparing genomic sequences is to align them—to map the letters of the sequences to each other. After an alignment is computed, visualization frameworks become essential to enable users to interact with the sequence and conservation data, especially in the context of longer DNA sequences or whole genomes. Visualization frameworks should be easy to understand by a biologist and provide insight into the mutations that a particular genomic locus has undergone.
The VISTA portal is a comprehensive comparative genomics resource that provides biomedical scientists with a single unified framework to generate and download multiple sequence alignments, visualize the results in the context of existing annotations and analyze comparative results in search for important sequence signals in alignments. The VISTA suite of programs has been in development and continued use since 2000 (1–4). It was originally developed for the alignment and comparative analysis of long genomic sequences and later was expanded to pair-wise and multiple alignment of vertebrate genomes. VISTA has popularized the visualization of the level of conservation in the format of a continuous curve based on the conservation in a sliding window. This concept proved to be extremely successful due to the easy interpretation of the resulting plots.
VISTA was built through a close collaboration between computational and biological scientists, resulting in a product that is robust, efficient and powerful, yet simple to use for a person without extensive computer experience, as is illustrated by more than 1000 citations to the various VISTA-associated tools (according to http://scholar.google.com).
In the last 2 years the VISTA portal has seen many significant improvements. In addition to updating the whole genome alignments, computed using recent assemblies of vertebrate, insect, plant and microbial genomes, we have added significant new functionality and resources to the Genome Browser and other tools, including:
A novel multiple whole genome alignment algorithm.
A new server for whole-genome alignment of bacterial genomes.
Base-pair level visualization ability within the VISTA browser.
Visual access to the results of the prediction of potential deleteriousness of non-synonymous Single Nucleotide Polymorphisms (SNPs) by the algorithm PolyPhen (5).
A novel conservation track, Rank-VISTA, to show the statistical significance of conserved regions computed by the Gumby algorithm (6).
Whole-genome rVISTA, that allows for evaluation of which conserved transcription factor binding sites (TFBS) are over-represented in a group of genes.
VISTA PORTAL
The suite of VISTA tools is accessible through the website http://genome.lbl.gov/vista. Currently it includes five servers for the analysis of user-submitted sequences: mVISTA that computes alignments of user-submitted sequences; wgVISTA to align whole genomes (up to 10 megabases in length); GenomeVISTA that aligns a user-submitted sequence to a selected genome assembly; rVISTA that searches for conserved TFBS; and Phylo-VISTA to analyze multiple DNA sequence alignments of different species while considering their phylogenic relationship. In addition, multiple whole-genome alignments of vertebrate, insect and plant species have been built using in-house algorithms and are publicly available for browsing and analysis. The portal provides access to the VISTA Genome Browser, the main visual interface for both the pre-computed whole genome alignments and alignments of user-submitted sequences. In the sections below we will discuss various algorithmic and functional improvements to the VISTA portal in the last 2 years.
Algorithmic improvements
Most of the algorithms for multiple alignment either rely on a reference genome, against which all of the other sequences are laid out, or require a one-to-one mapping, where each nucleotide of one genome is constrained to align to at most one place on the other genome (4,7–9). Both approaches have drawbacks for whole-genome comparisons. The first approach requires computation, storage and analysis of several alignments, one for each base species. The second approach fails to align any gene that has undergone duplications since the divergence of the species being compared. Additionally, ‘referenced’ alignments commonly fail to include elements conserved among some genomes, but missing in the base genome. We have developed and implemented a novel alignment algorithm that treats all genomes symmetrically.
Improved alignment pipeline
Initially our whole genome alignment pipeline used an alignment strategy where one genome was split up into contigs of about 250 kilobases (kb) (3,4). The potential orthologs for each contig were found in the second genome with the BLAT local aligner (12). This step was followed by a global alignment of two orthologous sequences. Although this approach produces a map that is more accurate within large syntenic blocks than an all-by-all local alignment, it has two main weaknesses: (i) small syntenic blocks, resulting from rearrangements within a larger region, may be missed; (ii) the initial arbitrary division of one genome into segments can split a syntenic region, making it difficult to map the region to its true orthologue.
To address these issues we have developed a ‘glocal’ alignment method, which treats the rearrangement events explicitly. There have been several algorithms that decide whether to accept or reject a local alignment based on other alignments near it, and thus allow for the direct treatment of the various rearrangement events. These include Shuffle-LAGAN (S-LAGAN, (10)), which currently serves as the underlying engine for our whole genome alignments. The pair-wise algorithm described below is based on a novel-chaining tool, called SuperMap. The multiple alignment algorithm is a progressive extention of the pair-wise one, where at every internal node we pick an ordering of the alignments that simplifies the next alignment that we will conduct. These algorithms will be discussed in detail in a separate publication.
Pair-wise alignment
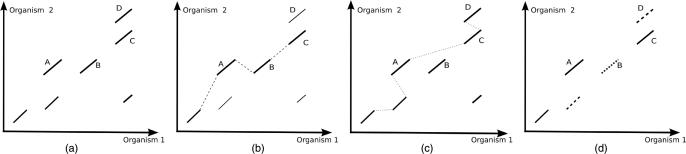
To align two genomes our algorithm uses a novel approach based on a reimplementation of the original S-LAGAN chaining algorithm (10,11) combined with a novel post-processing stage called SuperMap. The S-LAGAN chaining takes as input a set of local alignments between the two sequences generated by BLAT (12) or any other local aligner and returns the maximal scoring subset of these under certain gap criteria. This subset is called a 1-monotonic conservation map. In order to allow S-LAGAN to catch rearrangements, the map is allowed to be non-decreasing (monotonic) in only one sequence, without putting any restrictions on the second sequence. The 1-monotonic chain can capture all rearrangement events besides duplications in the second genome. In order to allow our alignments to incorporate these events we have introduced the novel SuperMap algorithm that takes two S-LAGAN outputs to make our algorithm symmetric. We run S-LAGAN twice, using each sequence as the base. This gives us three pieces of data: the original local alignments, which were common to the two runs of S-LAGAN, and two chains of these alignments, each corresponding to the S-LAGAN 1-monotonic maps. We then classify all local alignments as belonging to both chains, and consequently orthologous (best bi-directional hits) or being in only one chain, and hence a duplication (see Figure 1 for a graphical overview of the algorithm).
Figure 1.
SuperMap Algorithm. (a) Local alignment hits: regions A and B correspond to duplications in Organism 1; regions C and D correspond to duplications in Organism 2; (b) S-Lagan chain for Organism 1 as a base. Chain increases in direction of X-axis, but can jump up and down in Y-direction (Organism 2), region D is left out; (c) S-Lagan chain for Organism 2 as a base—chain increases in direction of Y-axis, region B is left out; (d) SuperMap output—combines regions of Figures b and c.
SuperMap has two advantages over regular S-LAGAN. One advantage is that it locates both regions of one-to-one similarity (those that were in both 1-monotonic chains) and likely duplications in both sequences (those in only one chain). Additionally, in case of transpositions, two of the pieces are no longer arbitrarily joined together.
Progressive multiple alignment
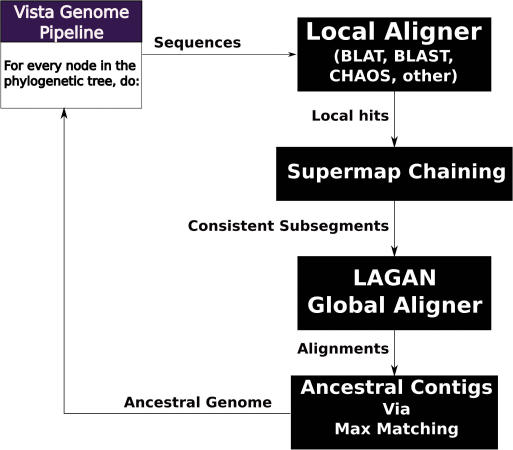
After the SuperMap algorithm is used to align the two pairs of sister taxa we use a progressive generalization of the pair-wise SuperMap algorithm to align all of the genomes, by following the species’ phylogenetic tree. After aligning two genomes, our algorithm joins together syntenic blocks (regions of genomes without rearrangements) based on their order in the outgroups (those sequences that will be aligned at a later stage: for example if we have aligned mouse with rat, then human, dog and chicken are all outgroups). We use an algorithm based on finding a maximum weighted matching in a graph, with the weights specified by the outgroup genomes, to order the individual alignment blocks in the order that will create the simplest alignment problem when we align the result to the outgroup. We then use the SuperMap based pair-wise alignment algorithm to align the alignments to each other using the regular LAGAN aligner (13). This algorithm is summarized in the flowchart in Figure 2.
Figure 2.
Multiple alignment with LAGAN in the VISTA Genome Pipeline (VGP). After running a local alignment program, SuperMap Chaining is used to identify all rearrangements. The resulting regions are aligned with LAGAN, and finally a maximum matching algorithm is used to predict ancestral contigs. These ancestral contigs are then used to align to outgroup genomes is the higher levels of the phylogenetic tree.
By picking an order of the syntenic blocks which is closest to the outgroup we facilitate the alignment of the more distant genomes. Our approach has several advantages over previous algorithms: (i) it does not assume a base genome, to which all other genomes are aligned, but creates a symmetric alignment equally valid for all genomes; (ii) it penalizes various rearrangement events based on an evolutionary tree, creating a set of alignments that mirror the evolutionary history of the sequences; and (iii) it is able to align short, low similarity syntenic blocks based on their adjacency to higher similarity areas even when there has been a rearrangement event between them.
wgVISTA: whole genome alignment for user's genomes
In order to allow our users to compare whole genomes using the whole genome alignment algorithms described above we have developed whole genome VISTA (wgVISTA), a tool which accepts sequences up to 10 megabasepairs in length, aligns them using our alignment pipeline and visualizes the results through the VISTA browser.
Visualization improvements
The VISTA Browser allows for the exploration of alignments and annotations of DNA sequences. It shows any number of alignments on a particular base genome and is scalable to the size of whole mammalian chromosomes. At the larger scale, visual presentation of rearrangements, inversions and gaps in the alignment are also available through the browser. Because all of our alignments are built in a symmetric fashion (see above section) the user may select any sequence or genome as the reference, and display the level of conservation between this reference and the sequences of other organisms. The browser has a number of options, such as zoom, extraction of a region to be displayed, user-defined parameters for conservation level and the selection of sequence elements for study. The VISTA Browser also gives access to the Text Browser that provides a user with all data related to alignments, analysis of conservation, and access to other resources. We have recently introduced two significant features into the VISTA browser that allow for a more detailed analysis of the areas of conservation detected in alignments.
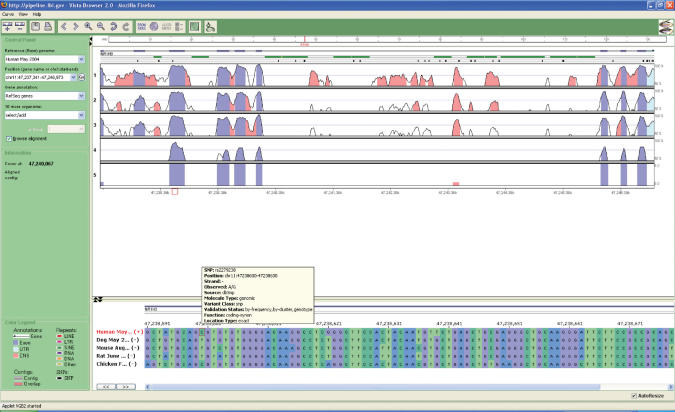
First, we have added a scrollable nucleotide-level alignment window. This window displays not only the details of the underlying pair-wise or multiple alignment, but also additional nucleotide-level annotation such as the SNP (Figure 3). Unlike the main VISTA window, the base-pair window does not have a selected continuous base sequence, but rather shows a real pair-wise or multiple alignment where a user can analyze gaps and substitutions in any sequence.
Figure 3.
VISTA Browser display of 9.6 kb fragment of NR1H3 gene on Chr. 11 of the human genome (hg17). VISTA plots for the five-way Human–Dog–Mouse–Rat–Chicken alignment are shown. Conserved sequences in VISTA (70%/100 bp cutoff) are colored according to the annotation (exons—dark blue, UTRs—turquoise and non-coding—pink). Rank-VISTA peaks identified by Gumby (P < 0.5) are shown as vertical bars following the same coloring convention. At the bottom of the window one can see the base-pair browser and SNP data [dbSNP annotation and PolyPhen (5) prediction of functionality for coding SNPs].
Second, we have introduced a plot to show the statistically significant conserved segments. The RankVISTA plotis a histogram-like plot where block width is proportional to the median conserved element length in human, and block area is proportional to the median of −log(P-value) (6). Block height thus represents the degree of evolutionary constraint at the base-pair level.
Finally, we have integrated the mVISTA server for user-submitted sequences with the VISTA Browser: when a user submits sequences to mVISTA, instead of just being e-mailed the VISTA results in a PDF document, we now make the alignment into a track on the VISTA browser, allowing the user to zoom in on a region of interest and view the detailed alignment in the nucleotide window.
New applications
One of the main emphases of our development in the last 2 years has been on better integration of the existing VISTA tools and of novel biological data into the VISTA portal. Several new applications developed in our group and by our collaborators have been integrated in to the VISTA portal, allowing biologists to easily access and visualize the results of these analyses.
Whole-genome rVISTA
Gene expression studies generate extensive lists of co-expressed genes, which can share regulatory factors that control their synchronous expression. We have developed a computational tool, called Whole-Genome rVISTA, designed to discover which conserved (between pairs of species) TFBS are over-represented in the upstream regions in a group of genes. This tool uses whole-genome alignments computed in our group, and TRANSFAC Professional from Biobase (14) with the MATCH program (15) to predict TFBS. The effectiveness of Whole Genome rVISTA was recently illustrated in a study of responsiveness to cAMP regulation (16). That expression study indicated that several circadian rhythm clock genes are induced by cAMP. We used Whole Genome rVISTA to scan 5Kb upstream of the transcription start site of the cAMP-regulated genes, and found that up-regulated genes contained more cAMP Response Elements (CRE) than all other genes on the array.
Gumby in RankVISTA
With more genomes available it has become essential to introduce new statistically motivated methods for conservation analysis that take into consideration neutral rates and phylogeny of the species. Gumby (6) makes no prior assumptions about evolutionary rates and requires no adjustment of parameters as the phylogenetic scope is varied from primates to vertebrates. Gumby uses a dynamically generated phylogenetic log-odds scoring scheme to identify local segments of any length that evolve slower than the background neutral rate, and ranks these conserved segments by P-value using the Karlin–Altschul statistic. This scoring technique demonstrated its efficiency in analyzing conservation both in evolutionary distant (17), and very close (6,18,19) species. Rank-VISTA plots of Gumby analysis allow the users to judge the statistical significance of any conserved regions and are available through VISTA Browser for genome-wide alignment of a number of genomes as well as for user-submitted mVISTA queries (Figure 3).
PolyPhen on the nucleotide alignment track
Poly/morphism/Phen/otyping (5) is a tool which predicts possible impact of an amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations. For each non-synonymous SNP in the dbSNP database (20) the VISTA Browser provides access to the results of the PolyPhen analysis of its deleteriousness.
FUTURE DIRECTIONS
The main emphasis of our future work within the VISTA portal will be on integration of additional data that is necessary for biological and medical researchers to carry out their analyses. We plan to integrate into our portal information about human variation, especially where it is known that some variation has a correlation with medical disorders. We will also continue to work on providing our users with a simple-to-use interfaces for browsing genomic data—we have been developing methodologies to display various evolutionary events in the context of the underlying phylogenetic trees (21,22) and expect to make similar improvements for visualizing rearrangements between the various genomes.
Finally, the new alignment pipeline implemented within the VISTA portal should be both flexible and powerful enough to analyze many of the genomes that are currently being sequenced. Consequently the majority of our alignment-related work in the near future will be on maintaining up-to-date versions of novel genomes, including low-coverage genomes that are currently being sequenced.
ACKNOWLEDGEMENTS
We are grateful to a large group of scientists and engineers who contributed to the VISTA project and whose names are listed on the VISTA Web site. Our special thanks to the biologists of the Genomics Division at LBNL for their help, support and critical comments. Research conducted at the E.O. Lawrence Berkeley National Laboratory was supported by grant HL066681 (L.A.P., I.D. and S.M.), Berkeley-PGA, under the Programs for Genomic Applications, funded by National Heart, Lung and Blood Institute and by HG003988 (L.A.P.) funded by National Human Genome Research Institute, and performed under Department of Energy Contract DE-AC02-05CH11231, University of California. Research in the Brudno laboratory was supported by the NSERC Discovery grant. Funding to pay the Open Access publication charges for this article was provided by grant HL066681 from NHLBI/NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 2.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brudno M, Poliakov A, Salamov A, Cooper GM, Sidow A, Rubin EM, Solovyev V, Batzoglou S, Dubchak I. Automated whole-genome multiple alignment of rat, mouse, and human. Genome Res. 2004;14:685–692. doi: 10.1101/gr.2067704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16:855–863. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bray N, Pachter L. MAVID: constrained ancestral alignment of multiple sequences. Genome Res. 2004;14:693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brudno M, Malde S, Poliakov A, Do CB, Couronne O, Dubchak I, Batzoglou S. Glocal alignment: finding rearrangements during alignment. Bioinformatics. 2003;19(Suppl. 1):i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- 11.Sundararajan M, Brudno M, Small K, Sidow A, Batzoglou S. WABI 2004, 4th Workshop on Algorithms in Bioinformatics. Norway: Bergen; 2004. Chaining algorithms for alignment of draft sequence. In: September 14–17, 2004. [Google Scholar]
- 12.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zambon AC, Zhang L, Minovitsky S, Kanter JR, Prabhakar S, Salomonis N, Vranizan K, Dubchak I, Conklin BR, et al. Gene expression patterns define key transcriptional events in cell-cycle regulation by cAMP and protein kinase A. Proc. Natl Acad. Sci. USA. 2005;102:8561–8566. doi: 10.1073/pnas.0503363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahituv N, Prabhakar S, Poulin F, Rubin EM, Couronne O. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum. Mol. Genet. 2005;14:3057–3063. doi: 10.1093/hmg/ddi338. [DOI] [PubMed] [Google Scholar]
- 18.Wang QF, Prabhakar S, Chanan S, Cheng JF, Boffelli D, Rubin EM. Detection of weakly conserved ancestral mammalian regulatory sequences by primate comparisons. Genome Biol. 2007;8:R1. doi: 10.1186/gb-2007-8-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang QF, Prabhakar S, Wang Q, Moses A, Chanan S, Brown M, Eisen M, Cheng JF, Rubin E, et al. Primate-specific evolution of an LDLR enhancer. Genome Biol. 2006;7:R68. doi: 10.1186/gb-2006-7-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007;35(Database issue):D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu S, Anderson I, Kunin V, Cipriano M, Minovitsky S, Weber G, Amenta N, Hamann B, Dubchak I. TreeQ-VISTA: an interactive tree visualization tool with functional annotation query capabilities. Bioinformatics. 2007;20:764–766. doi: 10.1093/bioinformatics/btl643. [DOI] [PubMed] [Google Scholar]
- 22.Shah N, Couronne O, Pennacchio LA, Brudno M, Batzoglou S, Bethel EW, Rubin EM, Hamann B, Dubchak I. Phylo-VISTA: interactive visualization of multiple DNA sequence alignments. Bioinformatics. 2004;20:636–643. doi: 10.1093/bioinformatics/btg459. [DOI] [PubMed] [Google Scholar]