Abstract
We developed OligoCalc as a web-accessible, client-based computational engine for reporting DNA and RNA single-stranded and double-stranded properties, including molecular weight, solution concentration, melting temperature, estimated absorbance coefficients, inter-molecular self-complementarity estimation and intra-molecular hairpin loop formation. OligoCalc has a familiar ‘calculator’ look and feel, making it readily understandable and usable. OligoCalc incorporates three common methods for calculating oligonucleotide-melting temperatures, including a nearest-neighbor thermodynamic model for melting temperature. Since it first came online in 1997, there have been more than 900 000 accesses of OligoCalc from nearly 200 000 distinct hosts, excluding search engines. OligoCalc is available at http://basic.northwestern.edu/biotools/OligoCalc.html, with links to the full source code, usage patterns and statistics at that link as well.
INTRODUCTION
Even prior to PCR, DNA oligonucleotides were used extensively in molecular biology as primers and as probes. With the availability of completely sequenced genomes, various genomic and array technologies including DNA microarrays and bead arrays have made oligonucleotides even more important reagents. OligoCalc provides a convenient web interface for calculating the physical properties of DNA and RNA oligonucleotides including melting temperature, molecular weight, %GC content and absorbance coefficient for a given oligonucleotide sequence. The recent interest and availability of biological applications for siRNAs has resulted in the addition of RNA oligonucleotides as common laboratory reagents, and OligoCalc can be used to calculate the properties of single-stranded and double-stranded RNA as well as DNA. OligoCalc provides the results of three common melting temperature calculations. Performing these calculations are all straightforward—enter the nucleotide sequence into a textbox, and hit return or click on the ‘Calculate’ button. In addition to these calculations, the user can enter absorbance readings to calculate the concentration of the oligonucleotide in ng/μmol and micrograms present in 1 ml of solution, and enter the predicted salt and/or primer concentrations in the final hybridization solution to more accurately predict the melting temperature. The user can also select options such as single- or double-stranded DNA and RNA molecules (ssDNA is the default), and the user can select from more than seventy 5′ and 3′ chemical modifications that refine the molecular weight and absorbance calculations for oligonucleotides with those modifications. Other options include the swapping of the entered sequence for its complement, submitting the sequence to the NCBI BLAST site, calculating self-complementarity between two identical oligonucleotide molecules, and calculating potential intra-molecular hairpin loop formation.
USING OLIGOCALC TO CALCULATE THE PROPERTIES OF OLIGONUCLEOTIDES
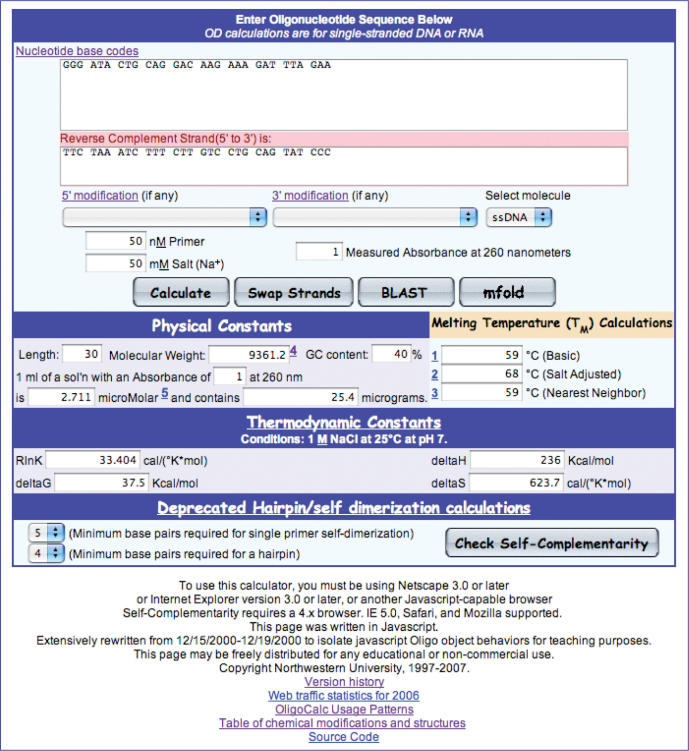
OligoCalc has a familiar ‘calculator’ interface and the basic properties can be calculated by pasting or entering the sequence followed by one of the following actions: clicking out of the sequence box, entering a ‘tab’, hitting ‘return’ or clicking ‘Calculate’. OligoCalc will use the currently entered sequence, selected options and entered conditions to calculate the length, molecular weight, estimated absorbance at 260 nm, the micromolar concentration and micrograms of oligonucleotide present in a 1 ml solution with an absorbance of one for the sequence entered. The calculator is available at the URL http://basic.northwestern.edu/biotools/OligoCalc.html and loading that URL in a browser will display the interface shown in Figure 1.
Figure 1.
Entry and main calculation screen for OligoCalc.
Once the user has entered a sequence, several additional options can be selected or entered. The absorbance at 260 nm (A260) can be entered for the oligonucleotide, and will result in the calculation of the micromolar concentration of oligonucleotide as well as the micrograms of the oligonucleotide present in a 1 ml solution with that absorbance. The millimolar concentration of salt [Na+] can be entered and will adjust the salt adjusted and nearest neighbor melting temperature calculations. The default value is 50 mM. The nanomolar concentration of primer in the hybridization solution can also be entered and will adjust the nearest neighbor melting temperature. The composition of the oligonucleotide (DNA or RNA, single-stranded or double-stranded) can be selected and will change many of the calculations, although the absorption coefficients are only accurate for single-stranded oligonucleotides. There are a number of 5′ and 3′ modifications that can be selected, and will change the molecular weight and in some cases the absorbance coefficient for the oligonucleotide. The entry of IUPAC codes are also supported (for instance W for A or T) and results in a range of values being reported for melting temperature, %GC content, molecular weight, concentration and micrograms present in a 1 ml solution with a A260 of 1, with the range representing the highest and lowest values possible for the set of possible oligonucleotides.
Clicking the ‘Swap Strands’ button swaps the entered strand for its reverse complement, and updates the properties of the oligonucleotide based on the new strand sequence. Note that if the ‘dsDNA’ or ‘dsRNA’ molecule type has been selected, swapping strands has no effect on the overall properties, since both strands are already taken into account by the calculations.
Clicking the ‘mfold button results in a new window that posts to the mfold web server (1,2) with likely hairpin and self-complementary areas highlighted.
A final option available is ‘BLAST’, which opens a new window and posts the sequence entered to the NCBI BLAST page and starts a blastn analysis of the entered sequence against the current set of non-redundant sequences (nr) available at the NCBI (3), with filtering for low-complexity sequences enabled.
There is also considerable documentation available, including a page of chemical modifications, including the chemical names for the common synonyms and links to the structures of the modifications, when available. This page is available at http://www.basic.northwestern.edu/biotools/OligoCalcModifications.html.
Available Calculations
Molecular weight calculations are based on the molecular weights available from Aldrich Chemicals, St. Louis, MO, USA. The absorbance coefficients and calculations are done as described in Molecular Cloning, a Lab Manual (4). The basic melting temperature calculation (5,6) is provided as a baseline for comparison, and is the least preferred method. The salt adjustment calculation is performed as described in Howley et al. (7) and the nearest neighbor thermodynamic calculations are done essentially as described by Breslauer et al. (8), but using the values published by Sugimoto et al. (9). RNA thermodynamic properties were taken from Xia et al. (10). The equations and the values used in all calculations are posted at the OligoCalc website, http://basic.northwestern.edu/biotools/.
Although OligoCalc is compatible with version 4 browsers (IE 4 and Netscape 4), browsers using either the Gecko or KHTML engines, or Internet Explorer 5.5 or higher are preferred. This includes Mozilla, Netscape 7, Camino, Safari, Konqueror or FireFox browsers. JavaScript must be enabled in the client browser for OligoCalc to work.
Available melting temperature method comparisons
The basic melting temperature is the least preferred method, however is perhaps the most often employed method for calculating melting temperature by bench scientists. OligoCalc was designed to give researchers an easy tool for finding and comparing melting temperatures using more accurate calculations. For oligonucleotides between 8 and 40 nt, the nearest neighbor method is the preferred method (8–12). Note that the equations and derived parameters were obtained using 14–20 mers, so this method is the most accurate for oligonucleotides of this length. A comparison of these data sets and recommendations were recently published (13) and implemented as a web server (14), and predominantly agree with the methods we have chosen. For longer sequences, or for oligonucleotides with base substitutions or modifications, the salt adjusted melting temperature calculation is the preferred method. Please note that these calculations are only estimates and many other factors can affect the melting temperature, including detergents, presence of other counter ions, solvents (ethanol for instance), formamide, etc.
ACKNOWLEDGEMENTS
The current utility of OligoCalc was aided by the initial prototype created by Eugene Buehler at the University of Pittsburgh Medical School which is still available at http://www.bioinformatics.org/JaMBW/3/1/9/index.html, the efforts of Qing Cao while she was a research associate at Northwestern, the monomer structures and molecular weights provided by Dr Bob Somers while at Glen Research Corporation. The initial requirements for OligoCalc came from a discussion with Dr Jim Kaput while he was the director of the Biotechnology Core Facility at Northwestern. The RNA calculations were a result of very helpful conversations with Dr Suzanne Kennedy at Qiagen, the fluorescent tags and options, along with the properties of the modified bases were requested and provided by Dr Regina Bichlmaier at metabion GmbH. I would also like to thank the reviewers, who made many substantial and insightful suggestions that dramatically improved this manuscript as well as the documentation and features available through OligoCalc. Finally, OligoCalc users have identified bugs and provided numerous suggestions and feature requests over the years. Thank you all. The Feinberg School of Medicine and the Robert H. Lurie Comprehensive Cancer Center of Northwestern University provided financial support for development and operation of OligoCalc. Funding to pay the Open Access publication charges for this article was provided by Northwestern University institutional sources.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 3.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 5.Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- 6.Wallace RB, Shaffer J, Murphy RF, Bonner J, Hirose T, Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979;6:3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howley PM, Israel MF, Law M-F, Martin MA. A rapid method for detecting and mapping homology between heterologous DNAs. Evaluation of polyomavirus genomes. J. Biol. Chem. 1979;254:4876–4883. [PubMed] [Google Scholar]
- 8.Breslauer KJ, Frank R, Blöcker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugimoto N, Nakano S, Yoneyama M, Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamics parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 11.SantaLucia J., Jr A unified view of polymer, dumbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owczarzy R, Vallone PM, Gallo FJ, Paner TM, Lane MJ, Benight AS. Predicting sequence-dependent melting stability of short duplex DNA oligomers. Biopolymers. 1997;44:217–239. doi: 10.1002/(SICI)1097-0282(1997)44:3<217::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 13.Panjkovich A, Melo F. Comparison of different melting temperature calculation methods for short DNA sequences. Bioinformatics. 2005;21:711–722. doi: 10.1093/bioinformatics/bti066. [DOI] [PubMed] [Google Scholar]
- 14.Panjkovich A, Norambuena T, Melo F. dnaMATE: a consensus melting temperature prediction server for short DNA sequences. Nucleic Acids Res. 2005;33:W570–W572. doi: 10.1093/nar/gki379. [DOI] [PMC free article] [PubMed] [Google Scholar]