Abstract
Wheat biologists face particular problems because of the lack of genomic sequence and the three homoeologous genomes which give rise to three very similar forms for many transcripts. However, over 1.3 million available public-domain Triticeae ESTs (of which ∼850 000 are wheat) and the full rice genomic sequence can be used to estimate likely transcript sequences present in any wheat cDNA sample to which PCR primers may then be designed. Wheat Estimated Transcript Server (WhETS) is designed to do this in a convenient form, and to provide information on the number of matching EST and high quality cDNA (hq-cDNA) sequences, tissue distribution and likely intron position inferred from rice. Triticeae EST and hq-cDNA sequences are mapped onto rice loci and stored in a database. The user selects a rice locus (directly or via Arabidopsis) and the matching Triticeae sequences are assembled according to user-defined filter and stringency settings. Assembly is achieved initially with the CAP3 program and then with a single nucleotide polymorphism (SNP)-analysis algorithm designed to separate homoeologues. Alignment of the resulting contigs and singlets against the rice template sequence is then displayed. Sequences and assembly details are available for download in fasta and ace formats, respectively. WhETS is accessible at http://www4.rothamsted.bbsrc.ac.uk/whets.
INTRODUCTION
Wheat is the most widely grown crop in the world with massive importance for human nutrition. However, genomics and DNA sequence analysis in wheat present particular problems. Cultivated wheat (Triticum aestivum) is an allohexaploid species with three homoeologous genomes (A, B, D), each comprising seven pairs of chromosomes. All three genomes are very large, so that together they contain about 30× as much DNA as rice and 6× as much as the human genome. Due to the technical difficulties, the complete genome sequence of wheat will not be available for several years at the earliest. However, there is a rich resource of wheat ESTs of which there are ∼850 000 and a further ∼500 000 from other Triticeae species in dbEST [(1); January 2007]. These can be mapped to the genes of rice as the most closely related fully sequenced genome (2). In this way, all the ESTs derived from the same transcript can be grouped and linked to information on the orthologues in rice and Arabidopsis. This procedure thus facilitates the application of knowledge gained in model species, particularly Arabidopsis, to wheat crops. The aim of Wheat Estimated Transcript Server (WhETS) is to flexibly allow the user to assemble Triticeae ESTs mapped to rice genes in this way and provide access to the results in a convenient form.
A common way to exploit ESTs is to use the pre-existing assemblies such as Unigene (3). However, because WhETS assembles related sequences in real time, the user can adjust the set of ESTs to be used, alter the stringency setting and view the affect of the changes on the assembly. Also, by anchoring the ESTs to rice loci, non-contiguous ESTs representing the same genes are automatically treated a part of the same set. The three very similar homoeologues of wheat genes are frequently all expressed (4), but assembly programs do not normally separate these so they are grouped together in contigs. These homoeologous sequences are best identified by analysis of shared SNPs such as can be achieved with SNPserver (5) which uses autoSNP (6) algorithms to separate alleles or homoeologues. A similar approach is included in WhETS to provide a best estimate of homoeologue-specific sequence. Aligning the Triticeae ESTs to rice has the additional advantage of being able to infer likely intron position which can be used to derive allelic markers, an approach taken in the USDA wheat SNP database (http://wheat.pw.usda.gov/SNP/new/index.shtml). Part of the aim of WhETS is therefore to bring together the useful features of Unigene, SNPserver and USDA SNP database into one site tailored specifically for wheat transcripts. However, it also has features unavailable elsewhere, such as the ability to display Triticeae EST distribution corresponding to a set of rice loci and the option to filter sequences according to library source tissue.
DESCRIPTION
Database
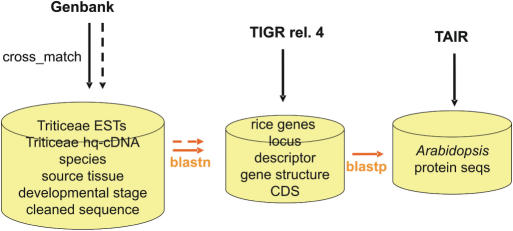
WhETS has a relational database containing sequences and annotation for all Triticeae EST and high quality cDNA (hq-cDNA) sequences from dbEST, coding sequences, annotation and intron positions for rice from The Institute for Genome Research (TIGR) rice pseudo molecule release 4 (7), and annotation for each locus from The Arabidopsis Information Resource (TAIR) version 6 (8) (Figure 1). EST sequences are first masked for vector contamination using the cross_match program (9). The WhETS database also contains the results of a blast (10) similarity searches: blastp of all the TIGR rice protein sequences against all the TAIR Arabidopsis proteins, and blastn of the Triticeae ESTs against the TIGR rice CDS. These tables contain the top scoring hits and any lower scoring hits with longer aligned regions, thus defining many-to-many relationships between Arabidopsis and rice genes and between rice genes and Triticeae sequences. The database is updated weekly by automated scripts which compare the contents with Triticeae entries in GenBank using Entrez utilities (11). Any missing entries are downloaded and any extra ones deleted. New sequences are subjected to a blastn search against the rice sequences and the sequences, and blast results are added to the WhETS database (Figure 1).
Figure 1.
WhETS database preparation steps. Dashed arrows indicate steps which are repeated in automatic weekly updates.
Real-time operation
The main part of WhETS requires TIGR rice loci identifiers. Users can start directly by supplying these as input, or they can start with a set of Arabidopsis AGI numbers or Triticeae accession numbers. WhETS will then retrieve all the matching rice loci for these. The user can then select filter settings for species, tissue and sequence type (EST or hq-cDNA). WhETS will then display the number and accessions of all the matching Triticeae sequences for each locus. The user can then select the locus for which they wish to obtain sequences for in the main part of WhETS.
When a single rice locus is selected, the user can again filter for species, tissue and sequence type. The sequences which pass this filter are assembled using the CAP3 program (12), and the resulting contigs are passed to an algorithm which analyses shared SNPs. If the contigs are found to contain groups which share more SNPs (i.e. base differences from the consensus) than a user-defined cut-off (default five SNPs per kb), these are split off into separate contigs. The CAP3 step tends to assemble paralogues which match the same rice locus into separate contigs, whereas the SNP analysis step is designed to separate homoeologues. However, by selecting higher stringency the user may also separate allelic forms. Conversely, in situations where there are relatively few ESTs it can be useful to assemble the sequences from wheat and related species with low stringency. WhETS also assembles the hq-cDNA sequences where present using a much higher base quality setting for the CAP3 program than used for ESTs. This has the effect that the consensus sequence of any contig will normally be the same as any hq-cDNA within it.
After assembly, the rice CDS is aligned to the contigs’ consensus and singlet sequences with blast and the results displayed using a modified version of a Perl script from the Korf et al. study (13). For contigs, links are supplied which open windows detailing all species, tissue, sequence type and cultivar of the constituent sequences. Singlets link out to the original NCBI entry. The main output for user downloading is a fasta file containing the rice template CDS, contigs’ consensus and singlet sequences. Additional details, such as intron positions are supplied in the descriptor fields of this file. Also available are other files, such as ace format files for each of the contigs containing all the information on the constituent sequences and their alignment, and a spreadsheet-compatible file containing details of all SNPs used to split contigs.
WhETS is implemented in MySQL (http://www.mysql.com/) and Perl using some Bioperl (14) modules. More details on allocation of blast hits within the WhETS database, strand of EST used and the algorithm for separating contigs into putative homoeologues are available in the Supplementary Data.
EXAMPLE OUTPUT
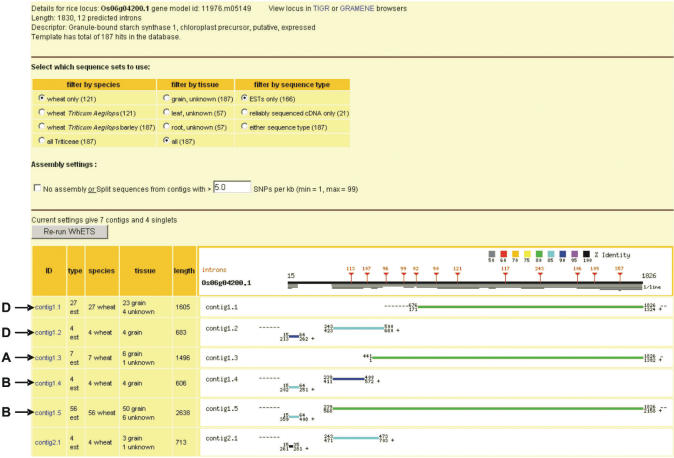
To test that WhETS correctly separates homoeologues, we examined the well-characterized WAXY locus, which encodes granule-bound starch synthase I. The three homoeologous forms are all sequenced, as are several allelic variants of these. As there are only a total of 2 715 wheat hq-cDNA sequences available, the normal use of WhETS is only with ESTs. We, therefore, ran WhETS with the orthologous rice locus Os06g04200 setting the filter to use ESTs and wheat sequences only. Figure 2 shows the output and how the resulting contigs match with the known homoeologues. From ESTs alone, WhETS correctly identifies the homoeologues and indicates the existence of a splice variant of the B homoeologue with a deletion in its 5′ UTR. Also shown (Figure 3) is the additional window detailing constituent sequences of one of the contigs.
Figure 2.
Output from WhETS for Os06g04200.1. The black line at the top corresponds to the rice gene CDS with intron position and size indicated as red vertical lines with triangles. Thin horizontal lines below this indicate the coverage of hits from the Triticeae sequences. The rows below show these hits, with blast HSPs for contigs and singlets shown as lines coloured according to percentage identity, and coordinates aligned to the rice template. The CAP3 step gives three contigs; one of these (contig 1) is then divided into five new contigs by the SNP analysis step (contigs 1.1, 1.2, etc.) The genome of origin (A, B, D) has been added to the screenshot according to 100% identity matches of the contig consensus to the known-homoeologue transcript sequences (exons of accessions AB019622, AB019623 and AB019624). Contigs 1.1 and 1.2 are not combined because of a lack of substantial overlap. Contigs 1.4 and 1.5 appear to be splice variants with an indel in the 5′UTR. Contigs 2 and 3 appear quite different and may be paralogues.
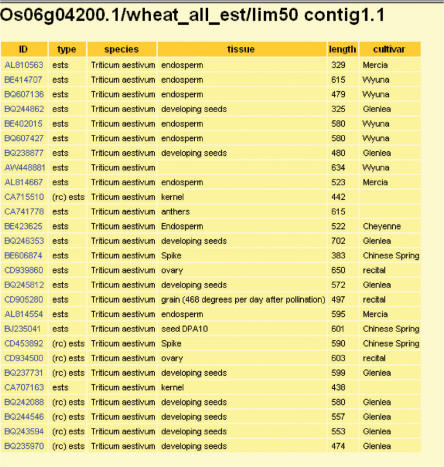
Figure 3.
Display window showing details for a contig which is opened by clicking on contig 1.1 link in Figure 2.
CONCLUSION
WhETS is designed to be a practical tool for wheat biologists to rapidly get the best estimate of transcript sequence for a target gene, supplemented with information on tissue distribution and likely gene structure. It is particularly aimed at producing wheat sequences from which to design PCR primers for cDNA templates.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENT
Rothamsted Research receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom. Funding to pay the Open Access publication charges for this article was provided by Rothamsted Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Boguski MS, Lowe TMJ, Tolstoshev CM. Dbest – Database for expressed sequence tags. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 2.Matsumoto T, Wu JZ, Kanamori H, Katayose Y, Fujisawa M, Namiki N, Mizuno H, Yamamoto K, Antonio BA, et al. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 3.Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mochida K, Yamazaki Y, Ogihara Y. Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol. Genet. Genomics. 2004;270:371–377. doi: 10.1007/s00438-003-0939-7. [DOI] [PubMed] [Google Scholar]
- 5.Savage D, Batley J, Erwin T, Logan E, Love CG, Lim GAC, Mongin E, Barker G, Spangenberg GC, et al. SNPServer: a real-time SNP discovery tool. Nucleic Acids Res. 2005;33:W493–W495. doi: 10.1093/nar/gki462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker G, Batley J, O'Sullivan H, Edwards KJ, Edwards D. Redundancy based detection of sequence polymorphisms in expressed sequence tag data using autoSNP. Bioinformatics. 2003;19:421–422. doi: 10.1093/bioinformatics/btf881. [DOI] [PubMed] [Google Scholar]
- 7.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee SY, Beavis W, Berardini TZ, Chen GH, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korf I, Yandell M, Bedell JA. BLAST. Sebastopol, USA: O’Reilly; 2003. [Google Scholar]
- 14.Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, et al. The bioperl toolkit: perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.