Abstract
Long terminal repeat retrotransposons (LTR elements) are ubiquitous eukaryotic transposable elements. They play important roles in the evolution of genes and genomes. Ever-growing amount of genomic sequences of many organisms present a great challenge to fast identifying them. That is the first and indispensable step to study their structure, distribution, functions and other biological impacts. However, until today, tools for efficient LTR retrotransposon discovery are very limited. Thus, we developed LTR_FINDER web server. Given DNA sequences, it predicts locations and structure of full-length LTR retrotransposons accurately by considering common structural features. LTR_FINDER is a system capable of scanning large-scale sequences rapidly and the first web server for ab initio LTR retrotransposon finding. We illustrate its usage and performance on the genome of Saccharomyces cerevisiae. The web server is freely accessible at http://tlife.fudan.edu.cn/ltr_finder/.
INTRODUCTION
LTR retrotransposons exist in all eukaryotic genomes (1–4) and are especially widespread in plants. They have been found to be the main components of large plant genomes (5–8). Dynamics of these elements are now regarded as an important force in genome and gene evolution. For example, their amplification and removal shape the organization and change the size of genomes (9,10); their transposition effects gene expression (11); and cases of gene movement via LTR retrotransposons were also reported recently (12). High throughput technologies for DNA sequencing are providing unprecedented chance to explore their functions and evolutionary impact on the basis of large-scale genetic information (13–16). It is urgent to develop efficient tools for locating these elements in rapidly deposited genomic sequences.
To date, most widely adopted methods of LTR retrotransposon identification in DNA sequences are based on alignment of known elements database to target genome. This class of methods can well detect elements in the database, but can hardly discover elements that is far related to or not in the database. On the other hand, analysis of many sequences of LTR elements in nearly 20 years revealed some structural features (signals) common in these elements, including Long Terminal Repeats (LTRs), Target Site Repeats (TSRs), Primer Binding Sites (PBSs), Polypurine Tract (PPT) and TG … CA box, as well as sites of Reverse Transcriptase (RT), Integrase (IN) and RNaseH (RH). These results have made ab initio computer discovery of LTR elements possible. However, tools for ab initio detection of LTR retrotransposons are still very limited: to the best of our knowledge, only two programs, LTR_STRUC (17) and LTR_par (18), have been reported, none of them being a web server.
We present here LTR_FINDER, a web server for efficient discovery of full-length LTR elements in large-scale DNA sequences. Considering the relationship between neighboring exactly matched sequence pairs, LTR_FINDER applies rapid algorithms to construct reliable LTRs and to predict accurate element boundaries through a multi-refinement process. Furthermore, it detects important enzyme domains to improve the confidence of predictions for autonomous elements. LTR_FINDER is freely available at http://tlife.fudan.edu.cn/ltr_finder/.
INPUT AND OUTPUT
User input
LTR_FINDER accepts DNA sequences file of FASTA or multi-FASTA format. Only the first ungapped string in the description line is recorded to identify the input sequence, and the rest of descriptions are ignored. In the sequence lines, Only A, C, G, T and N are allowed, and aligning an ‘N’ with any character is treated as a mismatch. Users are allowed to paste sequences in the ‘Sequence’ box, or upload a local file in the ‘File upload ’ box. The size of web uploading file should not exceed 50Mb. For users who need to scan very large size sequences, binary codes are available on request. When submitting a job, users can choose different parameters for different purposes. We explain some commonly used parameters here. The ‘tRNAs database’ of target species is for prediction of PBS. Because they are relatively conserved across organisms, tRNAs of a close related species can be used if those of the target species are not available. Since PBS is critical in deciding 3′boundaries of 5′LTRs, omitting this parameter will probably cause missing prediction. RT, IN and RH domains are important for an element to transpose. Occurrence of these sites adds weight of a candidate model to be a true autonomous element. If users choose domains in ‘Domain restriction’ options, only models containing selected ones are reported. ‘Extension cutoff’ controls if two neighboring exactly matched pairs should be joined into a longer one, that is, the regions covering them is regarded as a longer highly similar pair. ‘Reliable extension’ effects on identification of obscure overlapping elements. The higher the value is, the more models will be reported.
Program output
LTR_FINDER offers two types of output: full-output and summary-output. Full-output shows details of predictions, including LTRs sizes, element locations in the input sequence, similarity of two LTRs, sharpness (an index for boundary prediction reliability of LTR regions) and so on. Summary-output is extracted from full-output by omitting some detailed information. For each sequence, a diagram can be drawn simultaneously with either type of output. It visualizes location information of full-output. Users can obtain it by clicking on the ‘Output with figure’ button. The diagrams are convenient for human inspection and are very useful when analyzing potential overlapping elements: one can view the relative positions of signals inside LTR elements in details. In a diagram, two background colors, silver and white, are used to show sizes of objects. The program draws l pixels to represent l bases on the silver background while draws nlog(l) pixels to represent l bases on the white background, where n is a constant controlling overall size of the diagram. If users fill in the ‘Get result by e-mail’ box with a valid email address, the server will send the result instead of displaying it. The output file will be stored on the server for 3 days.
APPLICATION EXAMPLES
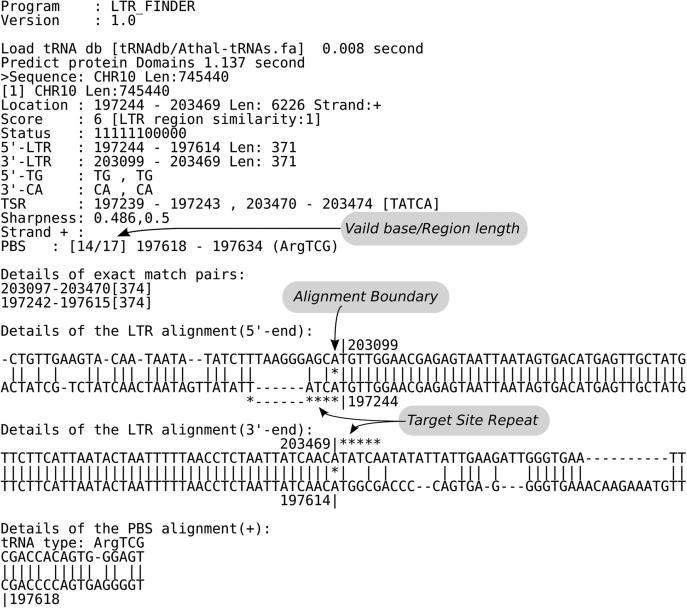
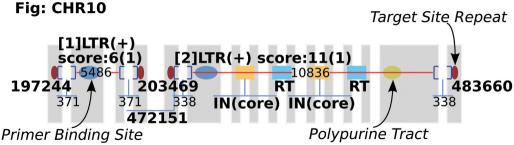
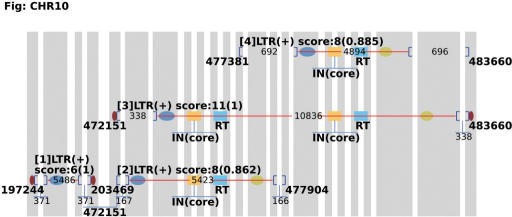
We describe an example of running LTR_FINDER on yeast chromosome 10 to show the usage of the server. Upload the sequence file, which can be obtained from Saccharomyces Genome Database (http://www.yeastgenome.org/). Here we use the version released on July 27, 1997 in order to compare the results with that described in (19), in which a standard benchmark of 50 full-length LTR retrotransposons on 16 yeast chromosomes were given. Using the default parameters, choosing ‘Saccharomyces cerevisiae tRNA database’ and ‘Output with figure’, we get the result as shown in Figures 1 and 2. Figure 1 gives a complete description of element 1 (pictures of the same element 1 appear in Figures 2 and 3). Explanation of the output items is given in the caption of Figure 1 and more information on output format can be found in documents on the webpage. The diagram of this run is shown in Figure 2. Yeast chromosome X contains a region where two tandem elements resulted from recombination. The program reports two sets of RTs and INs indicating the tandem structure (Figure 2, elements 2). A more sensitive search for overlapping elements by resetting ‘Reliable extension’ and ‘Sharpness lower threshold’ parameters reports the inserted LTR (Figure 3, element 3). Compared with the benchmark, locations of all elements are accurately predicted.
Figure 1.
LTR_FINDER sample output. ‘Status’ is an 11 bits binary string with each position indicating the occurrence of a certain signal. If a signal appears, the corresponding position is recorded ‘1’ and ‘0’ otherwise. From left to right, positions are as follows: [1] TG in 5′end of 5′LTR; [2] CA in 3′end of 5′LTR; [3] TG in 5′end of 3′LTR; [4] CA in 3′end of 3′LTR; [5] TSR; [6] PBS; [7] PPT; [8] RT; [9] IN(core); [10] IN(c-term) and [11] RH. ‘Score’ is an integer varying from 0 to 11. A detected signal adds 1 to its value.
Figure 2.
Diagram of two predicted elements with default parameters. Information of element 1 is shown in Figure 1. Element 2 is composed of two tandem LTR retrotransposons, which resulted from recombined insertion of a circular element. Two sets of enzyme domains are detected.
Figure 3.
Diagram of two tandem elements. Setting ‘Reliable extension’ to 0.95 and “Sharpness lower threshold’ to 0.2, the inserted element (element 3), its 5′LTR locating at 477837—478072, is reported.
Using the whole genome of yeast (∼12 Mb) as input, the web server implemented on a 600MHz PC took only 30 s, with RAM consumption <18 M. A total of 52 models were detected and all the 50 target elements were found. Among the test set, 48 were identified exactly, the remaining two predicted ones containing the targets with only 7 bp and 18 bp more in the 5′LTRs, respectively. The testing results gave no false negative and only two false positive reports, showing high speed, high sensitivity (100%) and specificity (96%).
LTR ELEMENT DISCOVERY STRATEGIES
LTR_FINDER identifies full-length LTR element models in genomic sequence in four main steps. The first step selects possible LTR pairs. In the beginning, LTR_FINDER searches for all exactly matched string pairs in the input sequence by a linear time suffix-array algorithm (20). Each pair, say a, is composed of two identical members: string located upstream (a5′) and downstream (a3′). Here upstream and downstream complies with that of the input sequence. Then it selects pairs of which distances between a5′ and a3′ as well as the overall sizes satisfy given restrictions. For each pair a and its downstream neighbor b, if the order of their locations in input sequence is 5′ a5′ … b5′ … a3′ … b3′ 3′, the regions [a5′,b5′] and [a3′,b3′] will be checked whether they should be regarded as a longer highly similar pair. Here ‘highly similar’ means that similarity between two members of the merged pair is greater than ‘Extension cutoff’). Calculation of the similarity involves in a global alignment of two regions: that inside two neighboring upstream strings and that inside two downstream strings. The pair keeps on extending until similarity between its members becomes less than ‘Extension cutoff’. Then it is recorded as an LTR candidate for further analysis. After that, Smith–Waterman algorithm is used to adjust the near-end regions of LTR candidates to get alignment boundaries. These boundaries are subject to re-adjustment again by TG … CA box and TSR supporting. At the end of this step, a set of regions in the input sequence is marked as possible loci for further verification. Secondly, LTR_FINDER tries to find signals in near-LTR regions inside these loci. The program detects PBS by aligning these regions to the 3′tail of tRNAs and PPT by counting purines in a 15 bp sliding window along these regions. This step produces reliable candidates. Additional validation comes from recognizing important enzyme domains. The program locates the most widely shared domain, RT, by first searching for its seven conserved subdomains, then chaining them together under distance restrictions using dynamic programming. This strategy is implemented to all six ORFs and is capable to detect RT domain even when there is a frame shift. For other protein domains such as IN and RH, it calls PS_SCAN (21) to find their locations and possible ORFs. At last, the program gathers information and reports possible LTR retrotransposon models at different confidence levels according to how many signals and domains they hit.
DISCUSSION
LTR_FINDER is the first web server devoted specially to full-length LTR retrotransposon discovery. It processes large-scale genomic sequences efficiently, which makes it applicable to rapid analysis of large genomes such as that of maize and wheat. A few improvements of the server are under way: (i) To make the interface more user-friendly, we plan to add buttons for automatic retrieval of sequences from GeneBank, EMBL and DDBJ by accession number to facilitate user input. (ii) LTR elements close to functional units (e.g. tRNAs, genes or centermeres) will be reported specially. The graphic output of the vicinity of LTR elements will be enhanced to reflect the local organization of functional units and LTR elements. (iii) It is also known that LTR elements may insert into internal regions of other elements to form nested structure. We expect LTR_FINDER to incorporate modules of finding nested elements.
ACKNOWLEDGEMENTS
The authors thank Bailin Hao for valuable comments and suggestions on the article, Xiaoli Shi for providing rice tRNA sequences and Heng Li for providing the linear-space pairwise alignment library. The authors are also grateful to all colleagues who helped testing the web server. Funding to pay the Open Access publication charges for this article was provided by Fudan University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ganko EW, Fielman KT, McDonald JF. Evolutionary history of Cer elements and their impact on the C. elegans genome. Genome Res. 2001;11:2066–2074. doi: 10.1101/gr.196201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc. Natl Acad. Sci. USA. 2003;100:6569–6574. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Voytas DF, Boeke JD. Yeast retrotransposon revealed. Nature. 1992;358:717. doi: 10.1038/358717a0. [DOI] [PubMed] [Google Scholar]
- 5.Flavell R. Repetitive DNA and chromosome evolution in plants. Phil. Trans. R. Soc. Lond. B. 1986;312:227–242. doi: 10.1098/rstb.1986.0004. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Bennetzen JL. Plant retrotransposons. Annu. Rev. Genet. 1999;33:479. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 7.Meyers BC, Tingey SV, Morgante M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat. Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 9.Vitte C, Panaud O. LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet. Genome Res. 2005;110:91–107. doi: 10.1159/000084941. [DOI] [PubMed] [Google Scholar]
- 10.Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- 12.Ma J, Devos KM, Bennetzen JL. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le QH, Wright S, Yu Z, Burea T. Transposon diversity in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy EM, Liu JD, Lizhi G, McDonald JF. Long terminal repeat retrotransposons of Oryza sativa. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-10-research0053. research0053.1–0053.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA. Structure and evolution of cereal genomes. Curr. Opin. Genet. Dev. 2003;13:644–650. doi: 10.1016/j.gde.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Wessler SR. Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc. Natl Acad. Sci. USA. 2004;101:5589–5594. doi: 10.1073/pnas.0401243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarthy EM, McDonald JF. LTR_STRUC: a novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19:362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 18.Kalyanaraman A, Aluru S. Efficient algorithms and software for detection of full-length LTR retrotransposons. J. Bioinformatics Comput. Biol. 2006;4:197–216. doi: 10.1142/s021972000600203x. [DOI] [PubMed] [Google Scholar]
- 19.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 20.Ko P, S. Aluru S. Space efficient linear time construction of suffix arrays. In: Baeza-Yates R, editor. Proceedings of the 14th Annual Symposium, Combinatorial Pattern Matching, LNCS; Springer-Verlag, Berlin, Heidelberg. 2003. pp. 200–210. [Google Scholar]
- 21.Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]