Abstract
Given the recent explosion of publications that employ microarray technology to monitor genome-wide expression and that correlate these expression changes to biological processes or to disease states, the determination of the transcriptional regulation of these co-expressed genes is the next major step toward deciphering the genetic network governing the pathway or disease under study. Although computational approaches have been proposed for this purpose, there is no integrated and user-friendly software application that allows experimental biologists to tackle this problem in higher eukaryotes. We have previously reported a systematic, statistical model of mammalian transcriptional regulatory sequence analysis. We have now made crucial extensions to this model and have developed a comprehensive, user-friendly web application suite termed the Promoter Analysis Pipeline (PAP). PAP is available at: http://bioinformatics.wustl.edu/webTools/portalModule/PromoterSearch.do
INTRODUCTION
It is becoming increasingly evident that the majority of human diseases are the product of multi-step processes each of which involves the complex interplay of a multitude of genes acting at different levels of the genetic program. Aberrant regulation at different stages of these processes may result in differential disease progression and treatment response and therefore may be used to define distinct disease subtypes (1,2). With the ultimate goal of improving diagnosis and treatment of human diseases, it is important to obtain a detailed view of the underlying molecular mechanisms of complex pathological processes. Such comprehensive and mechanistic understanding of the pathophysiology will offer insights into new therapeutic strategies and the development of new drug targets. Gene expression profiling has been used extensively in the study of complex diseases and to identify expression signatures that correlate with patient phenotypes (e.g. outcome or recurrence) (3,4). To better interpret these expression signatures, a reasonable next step is to take a systems biology approach to reconstruct the underlying genetic circuitry and regulatory program that distinguishes the aberrant cellular type from its normal state (5,6). Specifically, the identification of transcription regulators of co-expressed genes and the prediction of putative regulatory targets of one or more transcription factors will be crucial to deciphering the molecular aberrations underlying disease processes.
With the advancement of computational methods, researchers have attempted to uncover the transcriptional regulatory relationships in simple organisms (7–9). However, these methods have performed poorly when applied to higher species (10,11). Other software applications have utilized the idea of ‘phylogenetic footprinting’ to highlight regulatory elements that are conserved in multiple vertebrate species (12–16). However, analyses using such tools have been limited to the proximal promoter region of a gene (e.g. 10-kb upstream sequence). Studies have shown that functional regulatory elements may also be located in a distant upstream region (17) or within downstream intronic sequence (18,19). Thus, prediction of transcriptional regulation by these current programs may be incomplete. Moreover, no currently available tool is equipped with an integrated workbench, which incorporates a multitude of annotation data and facilitates the analysis of regulatory sequences of higher vertebrates efficiently and conveniently. Therefore, the prediction of the transcriptional regulatory mechanism underlying a given gene expression signature and the identification of potential transcription factor binding sites in human and higher animal model organisms remain a non-trivial task for experimental biologists. We have previously reported a systematic, statistical model for analyzing the transcriptional regulatory sequences in the proximal promoters in mammalian species (20). In this work, we have made crucial extensions to this model, namely the inclusion of four additional vertebrate genomes, inclusion of the sequence from the entire gene locus and the generation and usage of a non-redundant, high quality set of transcription factor binding profiles. These features are made available to bench scientists and translational researchers through the Promoter Analysis Pipeline (PAP), a comprehensive, user-friendly web application suite.
PAP is suitable for the identification of the potential transcriptional regulators of co-expressed genes and the identification of the potential regulatory targets of transcription factors. A typical PAP analysis includes input of a co-expressed gene cluster, identification of several high scoring transcription factors and visualization of the predicted transcription factor binding sites. The scoring scheme and the algorithm for the prediction of transcription factor binding have been described previously (20). More advanced features have been designed to assist users in the discovery of complex transcriptional regulatory network architectures. These include the identification of transcription factors that regulate a user-defined subset of the co-expressed genes or other genes in the genome that are regulated by the same transcription factors. Moreover, gene annotation and transcription factor information have been integrated seamlessly into PAP, and all the information generated in each step may be easily exported for further analyses or for publication.
MATERIALS AND METHODS
Data curation
In the previous version of PAP (version 1.0), genomic sequences were downloaded and processed, and transcription factor binding sites were pre-calculated and stored in order to predict higher mammalian transcriptional regulation in real time. Briefly, the 15-kb proximal promoter sequences of all the annotated genes in the human and mouse genomes were collected from GenBank. Repetitive elements in these sequences were masked. Orthologous gene pairs were determined using information from the HomoloGene, and conserved sequence regions between human and mouse orthologs were identified. All the evolutionarily conserved binding sites of characterized transcription factors in the non-coding sequences were then identified and used to calculate the binding probability score for each gene using a previously reported scoring function (Supplementary Data). Transcription factors that are mostly likely to regulate a co-expressed gene cluster could then be predicted by comparing these probability scores (20).
Although our previous version yielded promising results, it had a few limitations. While our model considered the evolutionary conservation of transcription factor binding sites, it included only two species, human and mouse. To improve the performance, three significant enhancements have now been included in a new version of PAP (version 2.0). First, we have now included four additional vertebrate genomes (rat, chimp, dog and chicken), draft sequences of which have recently been made available. This new dataset contains 21 237 human gene loci with annotated genomic locations, 2474 of which have alternatively spliced transcripts according to the annotation of the human genome build 35 (Supplementary Table 1, Supplementary Figure 1A and B). Of the six species, human has the highest content of repetitive elements in both the upstream intergenic region and the gene region (Supplementary Figure 1C and D). The sequence conservation of the gene locus and the intergenic sequences between human and other vertebrate species is shown in Supplementary Table 2. Using data from a total of six genomes, our model allows for the prediction of functional transcription factor binding sites across a wide spectrum of evolutionary conservation (e.g. conserved in all six species or conserved in only human and chimp). A gene may then be scored using the identified binding sites.
The second extension to our model is the inclusion of the entire genomic sequence of each gene's locus (i.e. exon and intronic sequence as well as the entire 5’ and 3’ intergenic regions). Our previous model only predicted conserved transcription factor binding sites within the 15-kb proximal promoter region of each gene. Although a large number of experimentally validated regulatory sequences are located very close to the transcription start sites, functional binding sites have been found in distant upstream regions or downstream intronic sequences. Therefore, computational models that only consider proximal promoter regions may miss bona fide sites and may be incomplete and biased. In our extended model, transcription factor binding sites are identified in two sets of sequences and are incorporated in the scoring system: those sites that are located within the 15-kb proximal region (the promoter region), and those that are located within the multi-species conserved sequences (MCSs) (21) in the whole gene locus. MCSs are the top 5% conserved sequences between human and rodents and are expected to cover most functional regulatory elements. Because MCSs are currently defined with respect to the human genome, they were mapped to the other five vertebrate genomes by using multiple sequence alignments (Supplementary Table 3).
A third improvement to our model is the generation and usage of a non-redundant, high-quality set of transcription factor binding profiles based on all the currently available profiles. Our previous model used a collection of transcription factor binding profiles in the TRANSFAC (22) and JASPAR (23) databases to predict the potential transcriptional regulators. While TRANSFAC curates a large amount of binding profiles, many profiles are redundant in the sense that they are very similar to each other and represent the same transcription factor. This generates redundancy in the analysis output and increases the difficulty in the interpretation of results. To overcome this problem, we have developed algorithms that use the program Matalign to compare and consolidate redundant binding profiles. Matalign calculates similarities between two input matrices and, when they are sufficiently similar, generates a new matrix by merging the two input matrices. Algorithm details are described in the supplementary information (Supplementary Data). In this process, 99 identical or highly similar matrices were eliminated (Supplementary Table 4). A total of 546 non-redundant weight matrices were generated, including nine new matrices created by merging multiple matrices (Supplementary Table 5). Transcription factor binding site prediction in PAP is conducted using these curated matrices.
Prediction testing
To test the improvement of transcription factor binding site prediction, results from PAP 2.0 were compared to PAP 1.0 by identifying binding sites in 14 muscle-specific genes reported previously (24). In PAP 2.0, not only were the number of predicted binding sites significantly reduced when additional genomes and non-redundant matrices were used, but additional highly conserved sites were also identified in introns and distant genomic regions (Supplementary Table 6). To determine the accuracy of prediction, 50 experimentally validated binding sites of muscle transcription factors were compared against those predicted by PAP. As additional genomes were included in the analysis, the positive predictive value increased almost 2-fold when using four genomes compared to the two available in PAP 1.0 (Supplementary Table 7). These results demonstrate that not only are additional binding sites predicted but that the accuracy of these predictions is greater in PAP 2.0.
User interface
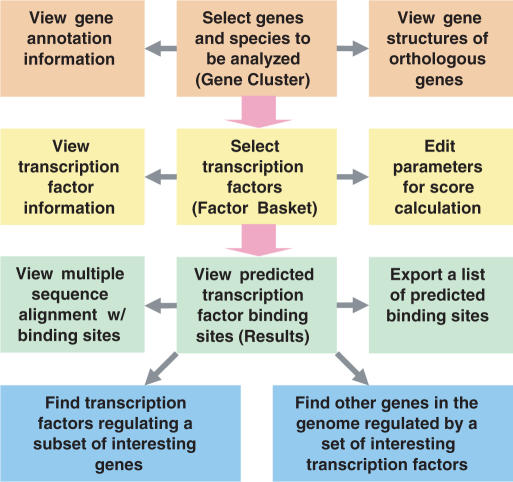
PAP is an interactive workbench which includes an array of analysis tools, packaged using a ‘wizard’ like interface. As the analysis proceeds, users are asked to provide input (e.g. input a set of genes for which binding sites are to be predicted) or make selections (e.g. set a threshold for selecting transcription factors), and the output is displayed at the end of each step. The data processing of the user interface is closely tied to a relational database, which stores all the pre-calculated results based on our computational model. The actual processing involves interpreting the input, querying the database and returning the results of the queries. The user interface of PAP consists of four major steps: gene selection, transcription factor selection, result visualization and advanced analyses (Figure 1). Each step includes comprehensive, online help documentation available at: http://bioinformatics.wustl.edu/webTools/portalModule/PromoterAnalysisHelp.do that describes the functionality and user input parameters in detail. In addition, movies demonstrating these steps are available on the website for novice users. Finally, all the features available through the web application are also exposed through a comprehensive and well-documented Application Programming Interface (API) available at http://bioinformatics.wustl.edu/webTools/portalModule/Api.do.
Figure 1.
Workflow of the user interface of PAP. A complete PAP analysis consists of four major steps: gene selection (orange), transcription factor selection (yellow), result visualization (green) and advanced analyses (blue). The major workflow of a typical analysis is shown in pink arrows. Links between pages are shown in gray arrows.
Gene selection
To start an analysis in PAP, the user is prompted to input a set of co-expressed genes, termed the Gene Cluster. Because gene annotation information is stored in our database, users may search using identifiers such as Entrez Gene IDs, gene symbols, RefSeq mRNA accession numbers, UniGene IDs and gene names. The user is also asked to determine the stringency of evolutionary conservation by selecting a subset of the six available species. This criterion plays a central role in analysis because only transcription factor binding sites that are conserved in the selected species will be considered and scored in the analysis. This setting may also be changed throughout the entire analysis, thus ensuring flexibility as the user proceeds through each step. After the user submits search criteria, PAP returns a paginated list of genes in the selected species satisfying the search criteria. For each gene, RefSeq mRNA accession numbers representing splice isoforms are listed. Because transcription factor binding sites are only searched in non-coding sequences, each transcript of a gene is scored separately in PAP. Therefore, the user must select which mRNA is to be used to represent a gene in this page. This feature of PAP allows for the investigation of differential regulation of alternatively spliced mRNAs. To aid the user in the selection of a representative transcript, exon–intron boundaries of all transcripts may also be visualized. Other functions in this page, including adding to and removing genes from the Gene Cluster, are described in detail in the online help document.
Transcription factor selection
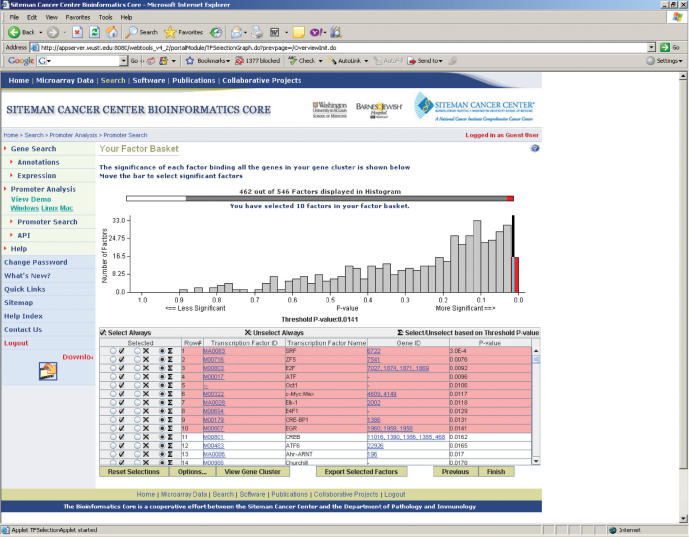
Using the genes in the Gene Cluster, PAP calculates the score of each transcription factor binding profile and displays the result in a table and histogram, which graphs the score distribution of all characterized transcription factors (Figure 2). All the non-redundant vertebrate matrices are included in this analysis regardless of the species of the genes in the Gene Cluster because orthologous transcription factors may bind to very similar sequences in different species. Significant transcription factors are then determined by a cutoff score, which may be adjusted by moving a vertical bar in the histogram. The transcription factors selected in this page, comprising the Factor Basket, are presumed to be transcriptional regulators of the genes in the Gene Cluster. Further analyses of PAP are based on transcription factors in the Factor Basket. This page is designed to provide multiple types of information which may assist the user in selecting potential transcription regulators of genes in the Gene Cluster. These include the score distribution of all the transcription factors, detailed information of each transcription factor binding profile and the annotation of genes encoding each transcription factor. Parameters of the transcription factor score calculation, such as the stringency of evolutionary conservation and the handling of genes whose orthologs are absent in some species, may be modified at this step.
Figure 2.
Screenshot of the Transcription Factor Selection page. The distribution of transcription factors as determined by their probability of regulating a set of ‘interesting’ genes is shown. The user may adjust the threshold to select high-scoring transcription factors.
Result visualization
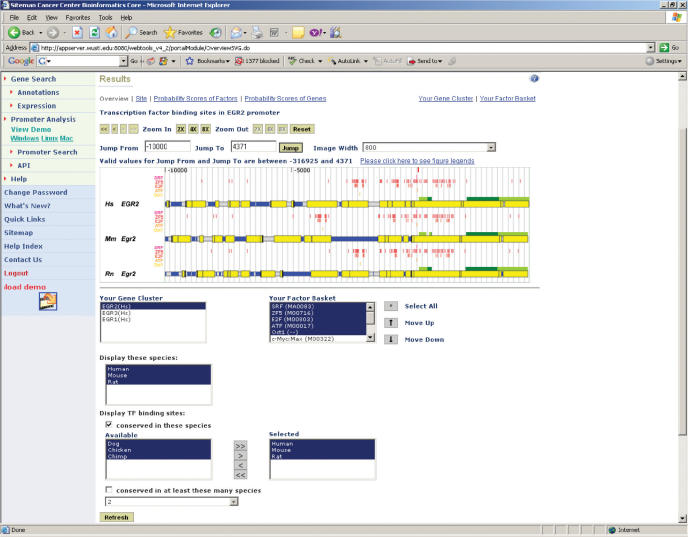
Predicted transcription factor binding sites in genes in the Gene Cluster may be viewed and examined in a sequence browser window in this step (Figure 3). The browser window, which initially displays the 15-kb proximal promoter region, may be used to display any sequence region within the gene locus using a set of navigation buttons that are similar in functionality to those found in the UCSC Genome Browser (25). A rich collection of sequence features, such as exons, introns, the transcription start site, repetitive elements, conserved sequence regions and transcription factor binding sites, are color-coded in the browser window. At this step, the user may again alter the stringency of evolutionary conservation of transcription factor binding sites. Other features of this page, such as changing the display order of transcription factors or viewing only a subset of orthologs, are designed to allow users to identify evolutionarily conserved transcription factor-binding site clusters (26,27). In addition, the alignment of a conserved sequence block annotated by transcription factor binding sites may be viewed by clicking on these elements in the sequence browser. In sum, the features available in this step permit end-users to identify relevant transcription factor binding sites and facilitate validation experiments (e.g. downloading alignments into a primer design application).
Figure 3.
Screenshot of the result overview page. Predicted transcription factor binding sites that are conserved in selected species are shown in a sequence browser. The user may alter the stringency of evolutionary conservation or view multiple sequence alignments annotated by binding sites in this page.
Advanced analyses
The first three steps of PAP support the major workflow of identifying transcriptional regulators of, and binding sites in a set of ‘interesting’ genes (i.e. genes in the Gene Cluster). Two natural extensions of this workflow include the identification of transcription factors that regulate a ‘subset’ of these genes and the comprehensive target prediction of regulators (i.e. transcription factors in the Factor Basket). These are described in more detail as follows. First, in the factor selection page, PAP calculates probability scores assuming all the genes in the gene cluster are regulated by the ‘same’ transcription factors. However, it is possible that only a subset of the input genes is regulated identically. Therefore, in the ‘Probability Scores of Factors page’ the user may select a subset of genes in the Gene Cluster and predict transcription factors that regulate this subset. Second, after having identified putative transcriptional regulators of a set of genes, a user may want to determine all transcriptional targets of those transcription factors in order to construct a comprehensive regulatory network. Thus, using the ‘Probability Scores of Genes’ page, the user may predict all the transcriptional regulatory targets of one or more transcription factors in the Factor Basket. Here, a score distribution of all the genes in the selected genome is calculated and displayed using the selected transcription factor binding profile(s). The genes that have been selected in the Gene Cluster are shown in the table and are highlighted in green in the histogram. The complete dataset of the probability scores of all the genes in the selected genome may also be exported in this page. In conclusion, these extensions permit users to discover complex regulatory patterns that may ultimately be used to construct comprehensive genetic networks.
Software design
PAP is a three-tier application composed of two major components. First, an integrated suite of PERL and C applications that runs on a Linux cluster is utilized to pre-process genomic sequences (e.g. masking repeats and creating multiple alignments), to identify transcription factor binding sites and to load this information into an Oracle 9i database. Second, the web application, developed using J2EE, Apache Struts, Java Applets and Scalable Vector Graphics, and deployed on Oracle Application Server10g, serves this pre-calculated data out to browser clients. The design of both components ensures that update of data (e.g. as new releases of genome sequences become available or as new versions of TRANSFAC and JASPAR are released) and addition of genomes (e.g. rhesus monkey and cow) requires no code change. Thus, PAP will be updated twice yearly and will include additional genomes as they are made available. Furthermore, an object-oriented design allows for the addition of new functionality (e.g. employment of new algorithms or computational models) with minimal effect to the original code base.
API
The PAP API, implemented in PERL, provides a rich set of functions that are exposed to software engineers and bioinformaticians through an XML-RPC service. The API, designed to insulate investigators from future database schema and algorithms changes, allows users to perform all the features available in the PAP web interface programmatically (e.g. retrieving the promoter sequences of a set of genes and predicting their transcriptional regulators). In addition, the API may be utilized to incorporate the functionality available in PAP into other software applications. Both example code of using the API and access to the XML-RPC service are available at: http://bioinformatics.wustl.edu/webTools/portalModule/Api.do.
CONCLUSION
In this work we present significant extensions to a previously developed regulatory sequence analysis model that are made available to the end-user through a web-based interface as well as an API. These extensions are crucial in that predictions are now more robust (i.e. conserved binding sites found across six genomes versus those found in only two genomes), reliable (i.e. due to our curation of binding profiles) and comprehensive (i.e. since the entire gene locus is included). The user-friendly web interface provides convenient access to the tools and data that we have developed and curated and permits experimental biologists and translational researchers to leverage computational biology approaches into their studies independently. Furthermore, the design of PAP ensures that it will continue to be useful as new data and algorithms become available in the future. For example, there are about 2000 human transcription factors curated in TRANSFAC, but only one-third of them have available weight matrices. Additional transcription factor binding profiles may be generated using pholygenetic footprinting approaches (28), and algorithms like PHYLONET (29), which make weight matrix model predictions. These are being considered for incorporation into future versions of PAP. In summary, the features in PAP facilitate the prediction of complex transcriptional regulatory mechanisms and the construction of genetic networks that may be dysregulated in complex diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Jeffrey Milbrandt for critical reading. G.D.S. is supported by the National Institutes of Health (NIH) grants HG00249 and GM63340. L.C. is supported by the National Institutes of Health (NIH) grants HG00249, GM63340, and K22LM008290. B.F. is supported by GM63340 and T32HG000045. R.N. is supported by K22LM008290. Funding to pay the Open Access publication charges for this article was provided by Washington University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- 2.Mooradian AD, Haas MJ, Wong NC. Transcriptional control of apolipoprotein A-I gene expression in diabetes. Diabetes. 2004;53:513–520. doi: 10.2337/diabetes.53.3.513. [DOI] [PubMed] [Google Scholar]
- 3.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 4.Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc. Natl Acad. Sci. USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat. Genet. 2005;37(Suppl. 1):S38–45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat. Genet. 2005;37(Suppl. 1):S31–S37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 7.Haverty PM, Hansen U, Weng Z. Computational inference of transcriptional regulatory networks from expression profiling and transcription factor binding site identification. Nucleic Acids Res. 2004;32:179–188. doi: 10.1093/nar/gkh183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouze P, Moreau Y. A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 2002;9:447–464. doi: 10.1089/10665270252935566. [DOI] [PubMed] [Google Scholar]
- 9.GuhaThakurta D, Palomar L, Stormo GD, Tedesco P, Johnson TE, Walker DW, Lithgow G, Kim S, Link CD. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res. 2002;12:701–712. doi: 10.1101/gr.228902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blais A, Dynlacht BD. Constructing transcriptional regulatory networks. Genes Dev. 2005;19:1499–1511. doi: 10.1101/gad.1325605. [DOI] [PubMed] [Google Scholar]
- 11.Siggia ED. Computational methods for transcriptional regulation. Curr. Opin. Genet. Dev. 2005;15:214–221. doi: 10.1016/j.gde.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aerts S, Van Loo P, Thijs G, Mayer H, de Martin R, Moreau Y, De Moor B. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005;33:W393–W396. doi: 10.1093/nar/gki354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lardenois A, Chalmel F, Bianchetti L, Sahel JA, Leveillard T, Poch O. PromAn: an integrated knowledge-based web server dedicated to promoter analysis. Nucleic Acids Res. 2006;34:W578–W583. doi: 10.1093/nar/gkl193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corcoran DL, Feingold E, Benos PV. FOOTER: a web tool for finding mammalian DNA regulatory regions using phylogenetic footprinting. Nucleic Acids Res. 2005;33:W442–W446. doi: 10.1093/nar/gki420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajiyama Y, Tian J, Locker J. Characterization of distant enhancers and promoters in the albumin-alpha-fetoprotein locus during active and silenced expression. J. Biol. Chem. 2006;281:30122–30131. doi: 10.1074/jbc.M603491200. [DOI] [PubMed] [Google Scholar]
- 18.Wong LH, Sim H, Chatterjee-Kishore M, Hatzinisiriou I, Devenish RJ, Stark G, Ralph SJ. Isolation and characterization of a human STAT1 gene regulatory element. Inducibility by interferon (IFN) types I and II and role of IFN regulatory factor-1. J. Biol. Chem. 2002;277:19408–19417. doi: 10.1074/jbc.M111302200. [DOI] [PubMed] [Google Scholar]
- 19.Mathew S, Mascareno E, Siddiqui MA. A ternary complex of transcription factors, Nished and NFATc4, and co-activator p300 bound to an intronic sequence, intronic regulatory element, is pivotal for the up-regulation of myosin light chain-2v gene in cardiac hypertrophy. J. Biol. Chem. 2004;279:41018–41027. doi: 10.1074/jbc.M403578200. [DOI] [PubMed] [Google Scholar]
- 20.Chang LW, Nagarajan R, Magee JA, Milbrandt J, Stormo GD. A systematic model to predict transcriptional regulatory mechanisms based on overrepresentation of transcription factor binding profiles. Genome Res. 2006;16:405–413. doi: 10.1101/gr.4303406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margulies EH, Blanchette M, Haussler D, Green ED. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 25.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, Papatsenko D, Small S. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc. Natl Acad. Sci. USA. 2005;102:4960–4965. doi: 10.1073/pnas.0500373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang T, Stormo GD. Identifying the conserved network of cis-regulatory sites of a eukaryotic genome. Proc. Natl Acad. Sci. USA. 2005;102:17400–17405. doi: 10.1073/pnas.0505147102. [DOI] [PMC free article] [PubMed] [Google Scholar]