Abstract
Although large-scale genetic association studies involving hundreds to thousands of SNPs have become feasible, the associated cost is substantial. Even with the increased efficiency introduced by the use of tagSNPs, researchers are often seeking ways to maximize resource utilization given a set of SNP-based gene-mapping goals. We have developed a web server named QuickSNP in order to provide cost-effective selection of SNPs, and to fill in some of the gaps in existing SNP selection tools. One useful feature of QuickSNP is the option to select only gene-centric SNPs from a chromosomal region in an automated fashion. Other useful features include automated selection of coding non-synonymous SNPs, SNP filtering based on inter-SNP distances and information regarding the availability of genotyping assays for SNPs and whether they are present on whole genome chips. The program produces user-friendly summary tables and results, and a link to a UCSC Genome Browser track illustrating the position of the selected tagSNPs in relation to genes and other genomic features. We hope the unique combination of features of this server will be useful for researchers aiming to select markers for their genotyping studies. The server is freely available and can be accessed at the URL http://bioinformoodics.jhmi.edu/quickSNP.pl.
INTRODUCTION
The biggest challenge in human genetics currently is to identify the genes whose alleles confer susceptibility to disease. It is believed that there will be many loci that increase the risk for each common disease (1). Since each causative gene may make only a very modest contribution to disease risk, identification of particular susceptibility variants becomes quite difficult. While genetic association studies have been used in gene mapping, their efficiency has been limited because they have typically assessed only one or a few genes at a time. The development of new SNP genotyping technologies, which can handle from dozens to hundreds of thousands of SNPs, and large numbers of samples, promises to accelerate gene mapping. Current platforms include the Illumina BeadArray and BeadChip systems (2,3), the Affymetrix GeneChip Mapping Arrays (4) and Applied Biosystem's TaqMan SNP Genotyping Assays (5). The newest technologies, while powerful, bring with them substantial costs, as they can involve as many as hundreds of millions of genotypes. For this reason, researchers have been trying to devise ways to maximize efficiency of resource utilization given a set of SNP-based gene-mapping goals.
It has been found that the pattern of linkage disequilibrium (LD) varies across the human genome and that there are discrete regions of high LD in the genome, called haplotype blocks (6). Most variation in populations can be characterized by a small number of common haplotypes. By selecting SNPs that uniquely identify or ‘tag’ these haplotypes, the number of markers and, hence, the cost of genotyping can be significantly reduced. The approach became more powerful with the availability of genetic data from the International HapMap Project (7), which contains genotype data for ∼4 million SNPs from each of four populations: Yoruba from Ibadan, Nigeria (YRI), Japanese from Tokyo (JPT), Chinese from Beijing (CHB) and United States residents with European ancestry (CEU).
Even with the increased efficiency introduced by tagSNPs, investigators are typically in the position of having to make strategic decisions about which set of tagSNPs to study. One strategy is to focus on those within genes, as these have the greatest likelihood of being functionally relevant or being in LD with those that are functional (8). Recently, a similar question was explored using empirical data from the HapMap-ENCODE project; tagSNPs chosen to capture common variation in exonic as well as evolutionarily conserved regions yielded genotype savings compared with a tagging approach that captured all common variation across the region (9). While the extent to which functionally important elements in the genome reside strictly within and near genes is not known, a gene-centric genotyping strategy may be a reasonable approach to searching for disease susceptibility alleles in the setting of limited resources.
The choice of SNPs for genetic association testing, thus, is a crucial step that will directly affect both the cost and the outcome of studies. Since the number of SNPs can range into the thousands, manual selection can be extremely time-consuming. There are some useful internet-based tools available for selection and prioritization of SNPs for genotyping. These include SNPper (10) (http://snpper.chip.org/), TAMAL (11) (http://neoref.ils.unc.edu/tamal/index.jsp), SNPSelector (12) (http://snpselector.duhs.duke.edu/hqsnp36.html), SNPHunter (13) (http://www.hsph.harvard.edu/ppg/software.htm), PupasView (14) (http://pupasview.bioinfo.cipf.es/) and tagger (15) (http://www.broad.mit.edu/mpg/tagger/). These programs have a variety of strengths as well as limitations. Among the gaps: most of them do not allow for automated selection of gene-based SNPs in a region, and none examines SNP coverage on genome-wide microarray SNP genotyping platforms.
We have developed a web server named QuickSNP to provide selection of tagSNPs in a chromosomal region, and to fill in some of the gaps in existing SNP selection tools. One useful feature of QuickSNP is the option to input the coordinates of a chromosomal region and have the program select SNPs, in an automated fashion, only from the genes within that region. Other useful features include automated selection of coding non-synonymous SNPs, SNP filtering based on inter-SNP distances, and reporting of whether SNPs have available assays or are present on whole genome chips.
There are several situations where we believe this tool will be particularly useful, including: (i) planning an LD-mapping study of a region de novo, where one has decided for any of a number of reasons to focus on genes and (ii) one is planning to obtain, or has obtained, data from a genome-wide association chip, and one wants to ‘fill in’ a particular region either because the chip scan produced a positive result or because of other information (e.g. a linkage peak or interest in a particular gene pathway), and one wants to find additional tagSNPs as well as coding non-synonymous SNPs in genes in the region.
MATERIALS AND METHODS
Implementation
QuickSNP utilizes Apache as its web server, and CGI (Common Gateway Interface) scripts are used to handle dataflow and validation to and from a dynamic HTML interface that utilizes cascading style sheet objects and integrated JavaScript. The data extraction and manipulation portion of the program is written in PERL (practical extraction and report language) modules and features two other programs embedded in the main code—Haploview, a freely available Java-based utility, and liftOver, a freely available Linux command-line application. QuickSNP is available at the URL http://bioinformoodics.jhmi.edu/quickSNP.pl. It is located on a cluster of processors running Linux OS at the Johns Hopkins McKusick-Nathans Institute for Genetic Medicine. All databases are locally downloaded and placed in the storage space of the Linux cluster. Files are not copied to a fileserver during user data uploads, but instead data is extracted dynamically from these files using CGI file handles, and thus information uploaded by users will not be retained on a file server.
Functionality
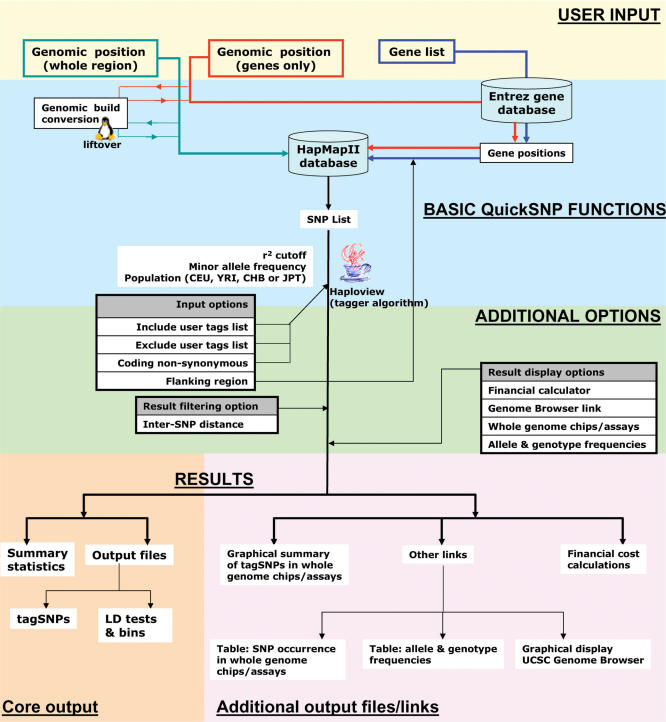
The basic function of QuickSNP is to generate a list of tagSNPs in a given chromosomal region, or for the genes in that region, or for any specified list of genes. For genomic position (whole region) searches, genotype data for SNPs lying in the region are extracted from the HapMap database (Figure 1). For genomic position (genes only) and gene name-based queries, gene coordinates are first extracted from the Entrez gene database and then SNP genotype data for those positions are extracted from HapMap. If the genomic position entered is not for NCBI build 35 (May 2004), it is first converted to that by the liftOver program. Also, genomic position is adjusted according to the length of flanking sequence used. The resulting SNP list is passed to the Haploview program, which generates tagSNPs based on the tagger algorithm (15) using the user-specified r2, minimum minor allele frequency and include/exclude tags specifications (if any).
Figure 1.
Schematic overview of the functioning of the QuickSNP web server.
There are categories of options that the user can select to obtain the best results from QuickSNP (see below). Input options like include/exclude tags and coding non-synonymous SNPs are used before tagSNP selection and they affect the list of SNPs that is used by Haploview for tagSNP selection. After selection of tagSNPs, the results can be further filtered by options such as removing SNPs lying too close (by a user-defined distance criterion). The user sees a results web page with two types of output. One is the core output consisting of a summary statistics table, a file displaying tagSNPs selected, and another file displaying pairwise LD tests as well as LD bins. The other type of output consists of additional information, including tables for genotype and allele frequencies, for the occurrence of SNPs in whole-genome chips and assays, and for the cost of genotyping. There is also a link to a graphical display of tagSNPs in the UCSC Genome Browser.
User interface
We have attempted to create a simplified user interface for QuickSNP so that it can be employed without the need for sophisticated computational skills. The input screen is divided into three main sections: input method, search conditions and additional options. The user may enter either genomic positions or gene names in the search window of the input method section. Multiple gene names can be entered by either typing in the corresponding window or uploading a file with a list of gene names. For the genomic position-based searches, users can further specify whether they wish to consider the whole region or just the genes within the specified region for tagSNP selection. The user is then required, in the search conditions section, to enter the desired r2, minor allele frequency and HapMap population.
To access the basic functionality of the server, the user need not consider the section containing additional options. However, depending on the study design, these options can enable more judicious and efficient selection of tagSNPs. For example, the user may want to include or exclude certain SNPs (based on availability of PCR primers or on past performance of genotyping assays). The user has the option to include flanking sequence around genes, reject SNPs that are too close to each other (because they are less likely to work with certain genotyping platforms), and force include coding non-synonymous SNPs, which can be identified and included automatically by QuickSNP through a search of the whole-genome coding SNP database.
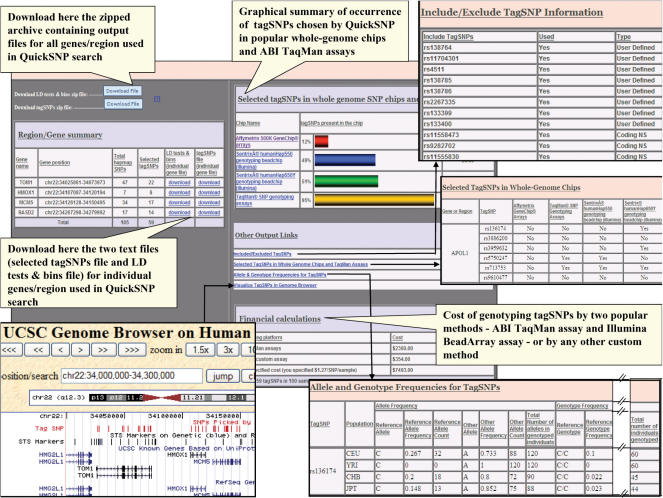
There are other result-related options that display various kinds of information for the chosen tagSNPs. These include the cost of genotyping using some popular methods, allele and genotype frequencies of tagSNPs in four HapMap populations, and occurrence of tagSNPs in available whole-genome chips and assays. For genomic position-based queries, the user also has the option to graphically visualize the tagSNPs in relation to genes, transcripts, conserved regions and other genomic features in that region using the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway). The results are generated in the form of a zipped archive containing multiple files (for multiple genes), as well as text files corresponding to an individual gene or region. A file containing a list of tagSNPs chosen and another file with details regarding LD tests and bins is generated for each gene/region. A summary table is also generated that displays the number of SNPs in the HapMap database for the gene/region queried and the eventual number of tagSNPs selected by QuickSNP for individual genes as well as the whole region. If include/exclude existing tags and/or coding non-synonymous SNPs were implemented in a QuickSNP search, an additional result table would be generated that lists the included or excluded SNPs, which of them were used in the tagSNP search (only those with genotype data in HapMap database can be used for tagging), and their type (user-specified include/exclude tags versus coding non-synonymous SNPs). Other results are also generated based on the additional options used for QuickSNP search (Figure 2).
Figure 2.
Snapshot of the results page generated by QuickSNP for a typical search with various components highlighted and explained.
There are three levels of help available to QuickSNP users: (a) QuickHelp, which can be accessed by clicking on the [?] symbol next to each option, and which briefly explains the purpose of that option; (b) frequently asked questions, which provides more detail and (c) direct contact with the authors, available by emailing us at QuickSNP@jhmi.edu.
VALIDATION AND USAGE EXAMPLE
We validated the core functionality of QuickSNP for various genes and genomic regions by comparing results from QuickSNP to those derived from a manual tagSNP selection using HapMap and the tagger algorithm in Haploview. Since many QuickSNP options and features are unique to this tool, they could not be compared to existing automated resources for tagSNP selection. For those cases, we manually performed steps of analyses for some of the options (for example, gene-based searches in a genomic region including coding non-synonymous SNPs), and compared the results with those generated in an automated manner by QuickSNP. The results generated by QuickSNP were always in agreement with those generated by the manual procedures.
We extensively used QuickSNP to select tagSNPs for a 6 Mb region on chromosome 17 that produced evidence for linkage to major depressive disorder in our Genetics of Recurrent Early Onset Depression (GenRED) collaborative project (16). Our aim was to select SNPs for an initial LD mapping association study of this region using the Illumina BeadStation custom genotyping platform. The region contained a total of ∼8000 HapMapII SNPs. Using criteria of r2 ⩾ 0.8 and MAF ⩾ 0.1, there were 1526 tagSNPs selected from across the full region and an additional 438 coding non-synonymous SNPs. Our project budget allowed us to study approximately 800 SNPs from the region in this initial experiment, so that excellent tagSNP coverage could be achieved if we focused on genes and their associated regulatory regions. We searched the region with QuickSNP using the genomic position, genes-only input method, force including coding non-synonymous and some previously genotyped SNPs, and rejecting SNPs that were closer than 60 bp. We performed various searches for different r2, MAF values and lengths of flanking region around genes. Table 1 shows the number of tagSNPs selected by QuickSNP using different combinations of parameters. We elected to genotype the 809 SNPs that resulted from tagging with r2 = 0.8, MAF ≥ 0.1 and a 5 kb flanking region on either side of each gene.
Table 1.
Number of tagSNPs selected for chromosome 17p 6.59–13 Mb region
| r2 | 0.8 | 0.9 | |||
|---|---|---|---|---|---|
| Minor allele freq. | 0.05 | 0.1 | 0.05 | 0.1 | |
| Flanking region around genes | 1 kb | 875 | 739 | 912 | 773 |
| 3 kb | 922 | 777 | 960 | 812 | |
| 5 kb | 960 | 809 | 999 | 845 |
Different lengths of flanking regions around genes and values of r2 and minor allele frequency cutoff were selected to generate this data.
SUMMARY
QuickSNP offers many useful features (see Table 2 for a comparison with other available programs):
Allows for a gene-centric approach to tagSNP selection;
Accepts multiple gene names as input;
Allows for automatic conversion of coordinates between different genome assemblies;
Provides the option to include flanking sequence around genes;
Provides the option to reject SNPs that are too closely spaced, since they are less likely to work in some genotyping platforms;
Calculates the cost for the genotyping study;
For the ‘include tag’ and ‘exclude tag’ options, predetermines which SNPs are present in the HapMap database for the given population, and implements inclusion or exclusion of only those (in other existing tools, the whole search aborts if any include/exclude tag is absent from the HapMap database);
Automatically includes coding non-synonymous SNPs in the region, if specified by the user;
Displays selected tagSNPs in the UCSC Genome Browser;
Reports allele and genotype frequencies for tagSNPs in different populations;
Reports the number of SNPs that have available assays or are present on whole genome chips provided by commercial genotyping platforms and
Provides a user-friendly summary table, and downloadable results files.
Table 2.
Comparison of QuickSNP features with those of comparable software programs
| SNPper | SNPSelector | SNPhunter | TAMAL | PUPASview | tagger | QuickSNP | ||
|---|---|---|---|---|---|---|---|---|
| INPUT- RELATED FEATURES | Gene name | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Chromosomal position | Yes | Yes | No | No | Yes | Yes | Yes | |
| Chromosomal band | Yes | No | No | No | Yes | No | No | |
| Batch query for gene names | Yes | Yes | No | Yes | Yes | No | Yes | |
| Conversion of coordinates between different genome assemblies | No | No | No | No | No | No | Yes | |
| Gene-centric tag selection in a chromosomal region | No | Yes | No | No | No | No | Yes | |
| FILTERING OPTIONS | MAF | No | Yes | No | No | No | Yes | Yes |
| r2 | No | Yes | No | No | No | Yes | Yes | |
| Option to force include/ exclude SNPs | No | Yes | No | No | No | Yes | Yes | |
| Selection of only relevant include/exclude SNPs for tagging* | No | No | No | No | No | No | Yes | |
| Include flanking region around genes | Yes | Yes | Yes | Yes | Yes | No | Yes | |
| Automatic inclusion of coding non-synonymous SNPs for tagging | No | No | No | No | No | No | Yes | |
| Spacing between SNPs | Yes | Yes | Yes | No | No | Yes | Yes | |
| OUTPUT-RELATED FEATURES | Allele and genotype frequencies for selected tagSNPs | No | No | No | No | Yes† | No | Yes |
| Financial cost of genotyping | No | No | No | No | No | No | Yes | |
| Occurrence of tagSNPs in popular whole genome chips and assays | No | No | No | No | No | No | Yes | |
| Representation of tag SNPs in UCSC genome browser | No | Yes | No | Yes | No | No | Yes |
*This option predetermines which of the include/exclude SNPs are present in HapMap database for the given population, and uses only those for tagging. If this criterion is not used, the whole search aborts if any one of the include/exclude tag is absent in the HapMap database.
†Only allele frequencies, but not genotype frequencies.
In the last few years, millions of new SNPs have been identified, and SNP genotyping technologies have developed rapidly. Investigators need to determine how to select SNPs for study in a chromosomal region in a manner that is efficient while still preserving power. There is a need for new tools, which can perform these functions in an automated manner. QuickSNP provides all of the basic SNP selection functions present in existing tools, while adding additional features.
FUTURE DIRECTIONS
At present, QuickSNP can handle regions as large as 5 Mb (for a genomic position-based search) or 40 genes (for a gene-name based search). In the future, we will attempt to optimize the algorithm and/or upgrade the hardware in order to increase this search limit. Since QuickSNP uses many public domain datasets, we will download and integrate the new updates as soon as they are released. An additional feature we are developing is the ability to assess the SNP coverage within genome-wide platforms for any given gene or genomic region. We will always welcome suggestions and bug reports by users, and will try to respond to these promptly.
ACKNOWLEDGEMENTS
We thank Drs Virginia Willour, Pamela Belmonte and Fernando Goes for testing the beta-version of QuickSNP and making helpful suggestions and we thank John Kloss for managing the Johns Hopkins McKusick-Nathans Institute for Genetic Medicine computational cluster where QuickSNP resides. Funding to pay the Open Access publication charges for this article was provided by National Institute of Mental Health, RO1 MH-059552.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 2.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;56–58(Suppl):60–61. [PubMed] [Google Scholar]
- 3.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, et al. Genotyping over 100 000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 5.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet. Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 6.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat. Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 7.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 9.Wiltshire S, de Bakker PI, Daly MJ. The value of gene-based selection of tag SNPs in genome-wide association studies. Eur. J. Hum. Genet. 2006;14:1209–1214. doi: 10.1038/sj.ejhg.5201678. [DOI] [PubMed] [Google Scholar]
- 10.Riva A, Kohane IS. SNPper: retrieval and analysis of human SNPs. Bioinformatics. 2002;18:1681–1685. doi: 10.1093/bioinformatics/18.12.1681. [DOI] [PubMed] [Google Scholar]
- 11.Hemminger BM, Saelim B, Sullivan PF. TAMAL: an integrated approach to choosing SNPs for genetic studies of human complex traits. Bioinformatics. 2006;22:626–627. doi: 10.1093/bioinformatics/btk025. [DOI] [PubMed] [Google Scholar]
- 12.Xu H, Gregory SG, Hauser ER, Stenger JE, Pericak-Vance MA, Vance JM, Zuchner S, Hauser MA. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics. 2005;21:4181–4186. doi: 10.1093/bioinformatics/bti682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Liu S, Niu T, Xu X. SNPHunter: a bioinformatic software for single nucleotide polymorphism data acquisition and management. BMC Bioinformatics. 2005;6:60. doi: 10.1186/1471-2105-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conde L, Vaquerizas JM, Ferrer-Costa C, de la Cruz X, Orozco M, Dopazo J. PupasView: a visual tool for selecting suitable SNPs, with putative pathological effect in genes, for genotyping purposes. Nucleic Acids Res. 2005;33:W501–W505. doi: 10.1093/nar/gki476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 16.Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe R, DePaulo JR, Jr, Knowles JA, Zubenko WN, Murphy-Eberenz K, et al. Genetics of recurrent early-onset depression: final genome scan report. Am. J. Psychiatry. 2007;164:248–258. doi: 10.1176/ajp.2007.164.2.248. [DOI] [PubMed] [Google Scholar]