Abstract
Real-world observable physical and chemical characteristics are increasingly being calculated from the 3D structures of biomolecules. Methods for calculating pKa values, binding constants of ligands, and changes in protein stability are readily available, but often the limiting step in computational biology is the conversion of PDB structures into formats ready for use with biomolecular simulation software. The continued sophistication and integration of biomolecular simulation methods for systems- and genome-wide studies requires a fast, robust, physically realistic and standardized protocol for preparing macromolecular structures for biophysical algorithms. As described previously, the PDB2PQR web server addresses this need for electrostatic field calculations (Dolinsky et al., Nucleic Acids Research, 32, W665–W667, 2004). Here we report the significantly expanded PDB2PQR that includes the following features: robust standalone command line support, improved pKa estimation via the PROPKA framework, ligand parameterization via PEOE_PB charge methodology, expanded set of force fields and easily incorporated user-defined parameters via XML input files, and improvement of atom addition and optimization code. These features are available through a new web interface (http://pdb2pqr.sourceforge.net/), which offers users a wide range of options for PDB file conversion, modification and parameterization.
INTRODUCTION
Due to the importance of electrostatic interactions in biomolecular systems, a variety of computational methods have been developed for evaluating electrostatic forces and energies [see (1–6) and references therein]. Typical computational electrostatics methods for biomolecular systems can be loosely grouped into two categories: ‘explicit solvent’ methods, which treat solvent molecules in full molecular detail, and ‘implicit solvent’ methods, which include solvent–solute interactions in averaged or continuum fashion. Implicit solvent methods are, by definition, limited in detail and therefore lack the atomic-scale accuracy of their explicit solvent counterparts. However, implicit solvent methods have gained increasing popularity, in part due to their elimination of the extensive sampling of solvent configurations required with explicit models (1,3–7).
The basic ingredients of an implicit solvent electrostatics calculation are environmental parameters such as temperature, solvent dielectric and ionic strength; biomolecular atomic coordinates; and parameters for atomic charges and radii. While the environmental parameters are relatively straightforward to specify, the remaining two ingredients can often be difficult to supply. In particular, most biomolecular structures in the Protein Data Bank (PDB) (8) do not contain hydrogen atoms, and many are also missing a fraction of the heavy atom coordinates. The addition of hydrogens and the reconstruction of these missing coordinates is not a trivial process; electrostatic properties obtained from the ‘repaired’ structures can often be very sensitive to the manner in which missing atoms are added and protonation states are assigned (9,10). Furthermore, inconsistent atomic nomenclature and other force field idiosyncrasies can often make the assignment of atomic charges and radii a cumbersome task. An additional obstacle to the use of PDB structures in electrostatics calculations and other biomolecular computational tasks is the accurate assignment of parameters to ‘non-standard’ residues and ligands.
Previously (9), we introduced the freely available PDB2PQR service (http://pdb2pqr.sf.net/), which was designed to facilitate the setup and execution of continuum electrostatics calculations from PDB data, particularly by non-experts. The original PDB2PQR server automated many of the common tasks of preparing structures for continuum electrostatics calculations, including adding a limited number of missing heavy atoms to biomolecular structures, estimating titration states and protonating biomolecules in a manner consistent with favorable hydrogen bonding, assigning charge and radius parameters from a variety of force fields, and finally generating ‘PQR’ output (a PDB-like format with the occupancy and temperature factor columns replaced with charge ‘Q’ and radius ‘R’, respectively) compatible with several popular computational biology electrostatics [APBS (10) and MEAD (11)], docking [AutoDock (12)], simulation [AMBER (13)] and visualization [VMD (14), PyMOL (15) and PMV (16)] packages. Since its inception, we have continued to expand the capabilities of the PDB2PQR server to address the challenges associated with ligand parameterization in PDB files and to include several new features.
METHODS
The PDB2PQR web service is driven by a modular, Python-based collection of routines, which provides considerable flexibility to the software and permits non-interactive, high-throughput usage. The service is available via a number of web mirrors listed at http://pdb2pqr.sf.net/. The source code is also available for download from this link, and due to the portability of Python, PDB2PQR can be executed on a wide range of platforms.
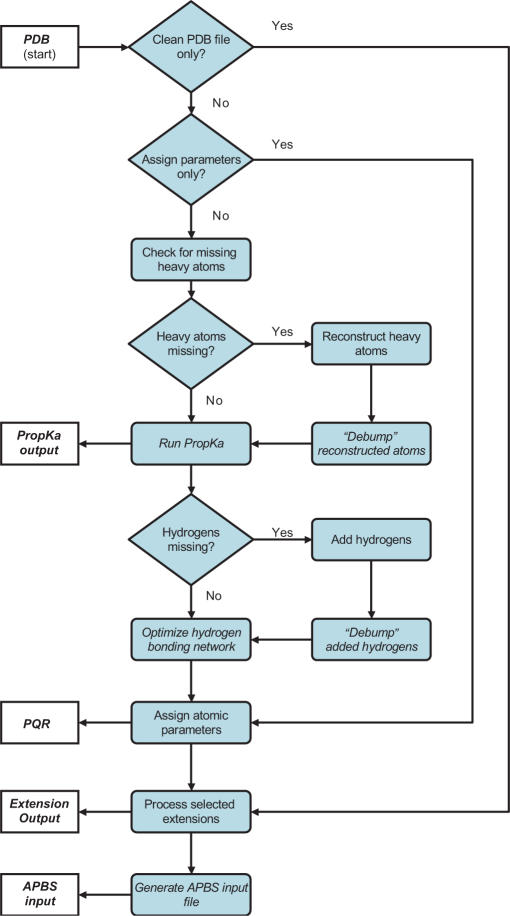
Figure 1 outlines the typical workflow of a PDB2PQR job and summarizes the features described in more detail below. The procedures for reconstruction of missing atoms, hydrogen optimization and APBS input generation were described previously (9) and are essentially unchanged in the current version of the software. Since their initial development, these atom reconstruction options have been greatly improved through a number of bug fixes and code optimization, robust support for separate biomolecular chains, and improved chain termini optimization. The following sections describe modified and new elements of the PDB2PQR pipeline.
Figure 1.
Flowchart demonstrating the sequence of operations performed by the pipeline. The process begins with an input PDB file and ends with a parameterized PQR file and, optionally, an APBS input file.
Titration state assignment by PROPKA
Protonation states for titratable protein groups are assigned by PROPKA 1.0 (http://propka.ki.ku.dk) (17). PROPKA utilizes a very fast empirical method to predict pKa values and is successful at predicting unusual pKa values. Recently, a comparative study of several protein pKa prediction methods showed that PROPKA was the most accurate method overall (18). PROPKA uses a heuristic method to compute the pKa perturbations due to desolvation, hydrogen bonding and charge–charge interactions. In the current version of PROPKA, contributions from nucleic acids as well as heteroatoms such as bound ions or ligands to the pKa values are not included. Note that, during the course of titration state assignment, PROPKA generates statistics on residue hydrogen bonding, location and solvent accessibility and Coulombic interactions. This information is available to users as a downloadable text file provided at the end of the PDB2PQR/PROPKA calculation.
Standard residue parameter assignment
PDB2PQR currently allows users to assign protein and (where available) nucleic acid parameters based on explicit solvent AMBER99 (19) and CHARMM27 (20) force fields, the PARSE continuum electrostatics force field (21), a Poisson–Boltzmann-optimized force field by Tan et al. (22), or user-defined force fields. User-defined parameters can be uploaded to the PDB2PQR server in a simple flat-file format described in the PDB2PQR user guide. Additionally, PDB2PQR output can be customized to include a variety of atom naming schemes, including AMBER99 (19), CHARMM22 (20), PARSE (21) and an internal naming scheme based on the IUPAC naming recommendations (23). This flexibility in nomenclature was included to facilitate import of PDB2PQR output into other modeling packages. Additionally, the web server provides a ‘map’ which is output at the end of every PDB2PQR calculation and presents a table of atoms’ name/number, residue name, chain name, AMBER atom type and CHARMM atom type to aid in the interpretation of parameter assignment and the development of user-defined charges and radii.
Ligand parameter assignment
The calculation of ligand charges necessitates detailed information on molecular structure and protonation states due to the large variation in the covalent structures of small-molecule protein ligands. The current version of PDB2PQR therefore requires the ligand structure, protonation state and formal charge to be specified by the user in the popular MOL2 (24) format. Ligand structures in MOL2 format are readily available from popular molecular modeling software and free web services such as PRODRG (25). Future versions of PDB2PQR will include a pdb2mol2 parser and automatic assignment of default ligand protonation states from a small-molecule pKa database.
The calculation of ligand charges in PDB2PQR is based on the partial equalization of orbital electronegativities (PEOE) procedure developed by Gasteiger and Marsili (26). In the PEOE procedure, orbital electronegativities χ are linked to partial atomic charges q by a polynomial expansion (χ = a +b·q + c·q2 + d·q3). The coefficients a, b, c and d were optimized by Gasteiger and Marsili using gas phase data on ionization potentials and electron affinities. We utilize a PEOE algorithm, which has been optimized by Czodrowski et al. to obtain better agreement between theoretical and experimental solvation energies for a set of small molecules including the polar amino acids (27). The resulting PEOE_PB charges have been tested for small-molecule complexes with trypsin, thrombin (28) and HIV protease (29), and have been found to give results that are in agreement with experimental values.
Post-processing
The current version of PDB2PQR supports an ‘extension’ directory for user-defined processing of PDB2PQR output. Such extensions might include alternative naming schemes, identification and parameterization of other molecule types, additional hydrogen bond processing, etc. The web servers listed at (http://pdb2pqr.sf.net/) provide only the default PDB2PQR functionality. However, it is straightforward for users to download the PDB2PQR software and setup their own web servers with additional functionality based on custom extensions.
CONCLUSIONS
We have described a number of new features for the free PDB2PQR web server, a service which helps users prepare molecular structures for further computational work by modeling missing atoms, assigning charges and titration states, and providing a mechanism for assignment of ligand parameters. Readers interested in these tasks might also be interested in other servers, which provide complementary services for biomolecular structure processing (30–32). Planned future developments for PDB2PQR include the construction of a pdb2mol2 parser to allow for the automatic parameterization of non-protein atoms, the correct treatment of protein post-translational modification, and the integration of a Poisson–Boltzmann continuum electrostatics-based pKa calculation algorithm into PDB2PQR. We anticipate that the PDB2PQR service will continue to be a helpful addition to the portfolio of tools available to the structural and computational biology communities.
ACKNOWLEDGEMENTS
N.A.B. and T.J.D. were supported by NIH grant GM069702 and the National Biomedical Computation resource (NIH P41 RR08605); J.H.J. and H.L. were supported by NSF grant MCB 0209941; J.H.J. gratefully acknowledges a Skou Fellowship from the Danish Natural Science Research Council; J.E.N. was supported by a Science Foundation Ireland PIYRA grant (04/YI1/M537); G.K. and P.C. were financially supported by the bilateral CERC3 program of CNRS and DFG (KL 1204/3). The authors would like to thank Andy McCammon for contributions to and support of early versions of the PDB2PQR effort. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM069702.
Conflict of interest statement. None declared.
REFERENCES
- 1.Baker NA. Improving implicit solvent simulations: a Poisson-centric view. Curr. Opin. Struct. Biol. 2005;15:137–143. doi: 10.1016/j.sbi.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Darden TA. In: Computational Biochemistry and Biophysics. Becker OM, MacKerell AD Jr, Roux B, Watanabe M, editors. New York: Marcel Dekker Inc; 2001. pp. 91–114. [Google Scholar]
- 3.Roux B. In: Computational Biochemistry and Biophysics. Becker OM, MacKerell AD Jr, Roux B, Watanabe M, editors. New York: Marcel Dekker; 2001. pp. 133–152. [Google Scholar]
- 4.Davis ME, McCammon JA. Electrostatics in biomolecular structure and dynamics. Chem. Rev. 1990;94:7684–7692. [Google Scholar]
- 5.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 6.Warshel A, Sharma PK, Kato M, Parson WW. Modeling electrostatic effects in proteins. Biochim. Biophys. Acta Proteins Proteomics. 2006;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Roux B, Simonson T. Implicit solvent models. Biophys. Chem. 1999;78:1–20. doi: 10.1016/s0301-4622(98)00226-9. [DOI] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup, execution, and analysis of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bashford D. In: Scientific Computing in Object-Oriented Parallel Environments. Ishikawa Y, Oldehoeft RR, Reynders JVW, Tholburn M, editors. Vol. 1343. Berlin: Springer; 1997. pp. 233–240. [Google Scholar]
- 12.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 13.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 15.DeLano WL. 2002. Palo Alto, CA, The PyMOL Molecular Graphics System. [Google Scholar]
- 16.Sanner MF. Python: a programming language for software integration and development. J. Mol. Graph. Mod. 1999;17:57–61. [PubMed] [Google Scholar]
- 17.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 18.Davies MN, Toseland CP, Moss DS, Flower DR. Benchmarking pKa prediction. BMC Biochemistry. 2006;7:18. doi: 10.1186/1471-2091-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 20.MacKerell AD, Jr, Bashford D, Bellot M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 21.Sitkoff D, Sharp KA, Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 1994;98:1978–1988. [Google Scholar]
- 22.Tan C, Yang L, Luo R. How well does Poisson–Boltzmann implicit solvent agree with explicit solvent? A quantitative analysis. J. Phys. Chem. B. 2006;110:18680–18687. doi: 10.1021/jp063479b. [DOI] [PubMed] [Google Scholar]
- 23.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol. 1998;280:933–952. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]
- 24.7.2 ed. St. Louis, MO: Tripos Inc.; 2006. SYBYL Molecular Modeling Software. ( http://www.tripos.com/mol2/mol2_format3.html) [Google Scholar]
- 25.van Aalten DMF, Bywater R, Findlay JBC, Hendlich M, Hooft RWW, Vriend G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 26.Gasteiger J, Marsili M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 27.Czodrowski P, Dramburg I, Sotriffer CA, Klebe G. Development, validation, and application of adapted PEOE charges to estimate pKa values of functional groups in protein-ligand complexes. Proteins. 2006;65:424–437. doi: 10.1002/prot.21110. [DOI] [PubMed] [Google Scholar]
- 28.Czodrowski P, Sotriffer CA, Klebe G. Protonation changes upon ligand binding to trypsin and thrombin: structural interpretation based on pKa calculations and ITC experiments. J. Mol. Biol. 2007;367:1347–1356. doi: 10.1016/j.jmb.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 29.Czodrowski P, Sotriffer CA, Klebe G. Atypical protonation states in the active site of HIV-1 protease: A computational study. J. Chem. Inform. Model. in press doi: 10.1021/ci600522c. [DOI] [PubMed] [Google Scholar]
- 30.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Jacobson MP, Zhu K, Zhao S, Friesner RA. Assignment of polar states for protein amino acid residues using a interaction cluster decomposition algorithm and its application to high resolution protein structure modeling. Proteins. 2007;66:824–837. doi: 10.1002/prot.21125. [DOI] [PubMed] [Google Scholar]
- 32.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]