Abstract
Terminal restriction fragment length polymorphism (T-RFLP) analysis is a widespread technique for rapidly fingerprinting microbial communities. Users of T-RFLP frequently overlook the resolving power of well-chosen restriction endonucleases and often fail to report how they chose their enzymes. REPK (Restriction Endonuclease Picker) assists in the rational choice of restriction endonucleases for T-RFLP by finding sets of four restriction endonucleases that together uniquely differentiate user-designated sequence groups. With REPK, users can provide their own sequences (of any gene, not just 16S rRNA), specify the taxonomic rank of interest and choose from a number of filtering options to further narrow down the enzyme selection. Bug tracking is provided, and the source code is open and accessible under the GNU Public License v.2, at http://code.google.com/p/repk. The web server is available without access restrictions at http://rocaplab.ocean.washington.edu/tools/repk.
INTRODUCTION
Terminal restriction fragment length polymorphism (T-RFLP) analysis is a microbial fingerprinting technique capable of discriminating microbial communities quickly and relatively inexpensively (1–3). T-RFLP is increasingly used in high-throughput studies of microbial communities in combination with or even in lieu of clone library analysis (4,5). Briefly, the method involves PCR amplification of a gene of interest (often 16S rRNA genes) with fluorescent dye-labeled primers, followed by multiple single restriction digests done in parallel. The resulting fragments are then separated by capillary electrophoresis with an internal size standard to determine the lengths of the terminal (fluorescently labeled) fragments. Each distinct terminal restriction fragment is considered an operational taxonomic unit (OTU), thus the choice of restriction enzymes can impact the number of OTUs observed in each sample and the calculation of diversity statistics.
When analyzing uncharacterized and very diverse bacterial communities, sufficient community discrimination can often be accomplished with multiple randomly-chosen tetrameric restriction enzymes (6). However, a brief review of the literature indicates that there is still no standard in even this simplified case. We examined 26 papers (1–5,7–26) that were published between 1997 and 2007 and used T-RFLP. Of those papers, 38% used universal bacterial primers combined with a single restriction enzyme, but the choice of enzyme was not consistent. MspI was used most frequently (four studies), followed by TaqI (two studies), and one study each used AluI, CfoI, HhaI and HaeIII. Overall, only three of the 26 papers included a rationalization of enzyme selection (1,2,17).
An alternate approach to T-RFLP can be taken if the microbial community has been characterized (by clone library analysis or by prediction from previous studies) or if a particular taxonomic group is being targeted with specific primers. In this case, a more reasoned choice of restriction enzymes can be conducted. In particular, specific species or microbial taxa of interest to the researcher—particularly closely related taxa that may share some restriction sites—can often be differentiated if the proper restriction enzymes are selected.
There are, however, few resources available to narrow down the selection process. Over 600 Type II restriction enzymes are commercially available, accounting for 262 distinct specificities (27). Existing computer programs for assisting in the choice of restriction enzymes include TAP-TRFLP (28), MiCA Enzyme Resolving Power Analysis (http://mica.ibest.uidaho.edu) and TRF-CUT (29). These programs perform in silico restriction digestions of a predefined sequence database or user-provided sequences, but these results must still be manually examined to determine which enzymes are best suited to discriminate that set of sequences. CLEAVER (30), a stand alone program, provides the above features as well as the ability to assign sequences to taxonomic groups at multiple levels and to search for enzymes that cut one group but not another group. However, it is limited to comparing only two groups at once. Restriction Endonuclease Picker (REPK) addresses this gap by finding enzymes that are able to discriminate an unlimited number of user-designated sequence groups on the basis of their terminal restriction fragment lengths. If no single enzyme can discriminate all groups, REPK reports sets of four restriction enzymes that together are able to differentiate the groups of interest. An important component of REPK is this ability to specify the taxonomic rank of sequences to be differentiated, which is particularly useful in the case where a diverse microbial community has been characterized by clone library analysis or there is an existing database of several subgroups of sequences that amplify with the same specific primers.
SITE USAGE AND EXAMPLES
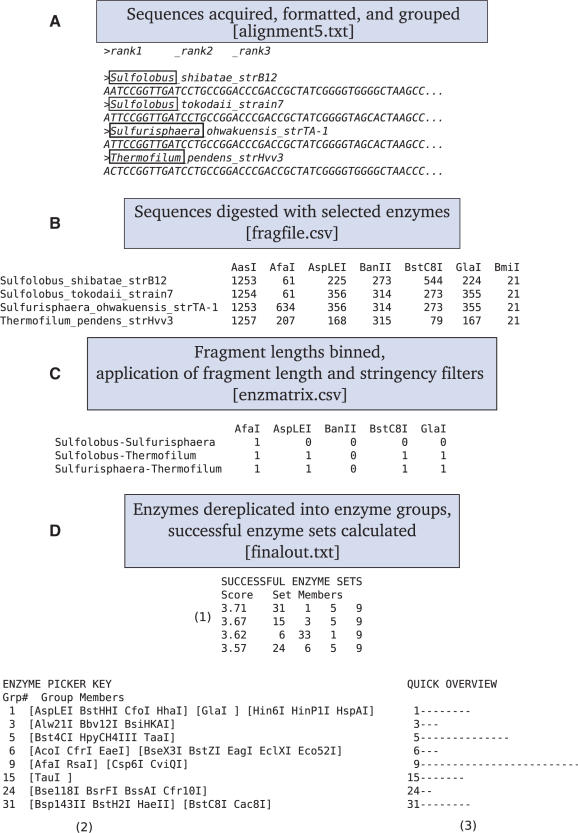
A complete manual and example input files are provided on the REPK website (http://rocaplab.ocean.washington.edu/tools/repk). The example shown in Figure 1 was prepared using REPK v. 1.0, with the following operating parameters (also the defaults): example sequence file (alignment5.txt), all commercially available Type IIP enzymes (REBASE Version 704), taxonomic rank = 1, cut-off = 5, min. fragment length = 75, max. fragment length = 900, stringency = ‘automatic’, max. missing groups = 0, max. matches returned = 100.
Figure 1.
Schematic summarizing the processing steps performed by REPK using program options detailed in the text, as well as subsets of example input and output files.
User input
The user must provide a trimmed FASTA-formatted file with nucleotide sequences beginning at the 5′-end of the labeled primer used for PCR amplification and ending at the 5′-end of the unlabeled primer. Sequence groups can be designated in the description line of the FASTA file, by using a delimiter to separate taxonomic rank terms or optionally taxonomic identifications can be prepended to the description line using an output file from RDP-Classifier (31). Figure 1A shows a subset of the example sequence file provided on the website, alignment5.txt. Sequence groups are separated by a single underscore, and in this example ‘taxonomic rank 1’ was chosen, corresponding to the genus of these Archaea.
A selectable list of commercially available enzymes from the latest REBASE database (27) is available and is automatically updated on the first day of each month. The enzymes available for selection include primarily Type IIP enzymes, which have symmetric recognition sequences and cleavage sites. Restriction enzymes of Type IIA (having asymmetric recognition sequences) and Type IIB (cleaving both sides of the recognition sequence on both strands) are at the present time not supported by REPK, although some are included in a separate enzyme file for advanced users willing to perform some manual processing. Users should be aware that some enzymes in the REBASE database may not be suitable for T-RFLP due to methylation specificities or requirements for multiple restriction sites to be present for effective digestion. Finally, users can define their own custom enzymes if they are not included in the standard list. The default (all standard enzymes) was used for the example in Figure 1. For computational efficiency isoschizomers are grouped by cleavage site.
The final output is refined by setting several options. Some of these, the minimum and maximum allowable fragment lengths and the maximum difference in size between two fragments that will still be considered the ‘same’ fragment, will be dependent on the specifications and resolving power of particular capillary electrophoresis systems. Users can also set the minimum threshold for the number of groups each enzyme must be able to discriminate on its own (the enzyme stringency), and the number of groups allowed to remain undifferentiated in the case that no ‘perfect’ enzyme groups are discovered.
Program operations
Sequences are first digested in both orientations by all selected enzymes to find the shortest labeled restriction fragment; these lengths are output as a table (and a downloadable tab-delimited text file, fragfile.csv), a subset of which is shown in Figure 1B. In this example, the sequences were cut by every enzyme except AasI, which resulted in full-length fragments.
Next, all terminal fragment lengths are binned within the chosen cut-off (here 5 bp) and a binary matrix of pairwise group differentiations is created. Bins containing a single sequence group yield a ‘1’, while bins containing more than one sequence group yield a ‘0’, indicating no differentiation between those groups. In the example in Figure 1, BanII failed to distinguish between sequence groups Sulfurisphaera and Thermofilum because the difference between their fragment lengths (1 bp) was less than the chosen cutoff of 5 bp (Figure 1B). However, AspLEI did distinguish between those groups because the difference in fragment lengths was 188 bp. It is not necessary for sequences from the same sequence group to have similar fragment lengths (e.g. Sulfolobus). Fragment lengths outside the boundaries set by the minimum and maximum fragment length options are binned together without regard for their actual lengths, decreasing the number of sequence groups discriminated by those enzymes (e.g. BmiI). The enzyme stringency filter is then applied to this matrix, allowing only enzymes that discriminate at least the specified fraction of sequence groups to proceed. The passing enzymes are output as a table (and a downloadable tab-delimited text file, enzmatrix.csv), a subset of which is shown in Figure 1C.
For computational efficiency, the enzymes are then sorted into ‘enzyme bins’ that produce identical differentiation patterns, although they may not produce the same terminal fragment lengths. In this example, neoschizomers AspLEI and GlaI produce different fragment lengths but the same differentiation pattern so they were grouped together for the final analysis. It is important to note that the enzyme bins are dependent on the particular sequence file and taxonomic rank selected for the analysis. That is, two enzymes may have equal discriminatory power for a particular set of sequence groups but for a different set of sequences, one enzyme may be much better and the two enzymes would be placed in the same bin in the first but not the second case.
Finally, groups of four enzymes (a ‘set’) are logically summed (e.g. 101 + 011 = 111) to determine the coverage of the set, i.e. the number of sequence groups discriminated by the enzymes in the set. If this number is greater than the total number of sequence groups (less than the max. missing groups, here 0) then the set is saved. A score is calculated for each saved set and all saved sets are sorted before the highest-scoring sets are output to a text file, finalout.txt, a subset of which is shown in Figure 1D. If more than 10 000 sets are found and the enzyme stringency is set to ‘automatic’, it is incremented by 10% (decreasing the number of passing enzymes and thus enzyme sets) and the analysis is repeated. The final output reports and summarizes those enzyme sets that best discriminated the sequence groups.
The final output consists of three parts: ‘successful enzyme sets’, ‘enzyme picker key’, and ‘quick overview’. The successful enzyme sets (Figure 1D.1) consist of a list of enzyme groups in each set, and a score indicating the frequency with which each set discriminated the sequence groups. A perfect enzyme (one that discriminates 100% of the sequence groups) contributes a score of 1, so four perfect enzymes would produce the maximum score of 4. The enzyme picker key (Figure 1D.2) lists the members of each enzyme group, with neoschizomers separated by brackets. Each member of an enzyme group produces the same sequence group differentiation pattern but may differ in recognition site, terminal fragment lengths, etc. The quick overview (Figure 1D.3) histogram summarizes the frequency with which each enzyme group appears in the printed results.
After submission the program generally takes less than 1 min to complete, depending most heavily on the number of sequence groups, the number of enzymes selected and the server load, respectively. The final choice of restriction enzymes is left to the researcher, and is likely to be based on practical factors such as cost, availability, reaction conditions, methylation sensitivity or requirements, star activity and other specifics that are detailed at REBASE. An online manual detailing usage and options, bug tracking and the source code (open and accessible under the GNU Public License v.2) are available at http://code.google.com/p/repk.
CONCLUSIONS
We found that researchers often failed to report their rationale in choosing a particular set of restriction enzymes for T-RFLP analysis, yet this choice is crucial for resolving the microbial community and interpreting the results. We provide REPK in the hope that it will allow microbial ecologists to maximize their ability to discriminate terminal restriction fragments obtained during T-RFLP and thereby take greater advantage of this powerful community fingerprinting technique.
ACKNOWLEDGEMENTS
R.E.C. was supported by NSF award OPP-0327244 and Washington Sea Grant award to J.W. Deming. G.R. was supported by NSF awards OCE-0822026 and OCE-0352190. Funding to pay the Open Access publication charges for this article was provided by XXX.
Conflict of interest statement. None declared.
REFERENCES
- 1.Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osborn AM, Moore ER, Timmis KN. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2000;2:39–50. doi: 10.1046/j.1462-2920.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- 3.Blackwood CB, Marsh T, Kim S-H, Paul EA. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 2003;69:926–932. doi: 10.1128/AEM.69.2.926-932.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tom-Petersen A, Leser TD, Marsh TL, Nybroe O. Effects of copper amendment on the bacterial community in agricultural soil analyzed by the T-RFLP technique. FEMS Microbiol. Ecol. 2003;46:53–62. doi: 10.1016/S0168-6496(03)00192-2. [DOI] [PubMed] [Google Scholar]
- 5.Moss JA, Nocker A, Lepo JE, Snyder RA. Stability and change in estuarine biofilm bacterial community diversity. Appl. Environ. Microbiol. 2006;72:5679–5688. doi: 10.1128/AEM.02773-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engebretson JJ, Moyer CL. Fidelity of select restriction endonucleases in determining microbial diversity by terminal-restriction fragment length polymorphism. Appl. Environ. Microbiol. 2003;69:4823–4829. doi: 10.1128/AEM.69.8.4823-4829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin KJ, Lukow T, Stubner S, Conrad R. Structure and function of the methanogenic archaeal community in stable cellulose-degrading enrichment cultures at two different temperatures (15 and 30 degrees C) FEMS Microbiol. Ecol. 1999;30:313–326. doi: 10.1111/j.1574-6941.1999.tb00659.x. [DOI] [PubMed] [Google Scholar]
- 8.Dunbar J, Ticknor LO, Kuske CR. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 2000;66:2943–2950. doi: 10.1128/aem.66.7.2943-2950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urakawa H, Yoshida T, Nishimura M, Ohwada K. Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ. Microbiol. 2000;2:542–554. doi: 10.1046/j.1462-2920.2000.00137.x. [DOI] [PubMed] [Google Scholar]
- 10.Stepanauskas R, Moran MA, Bergamaschi BA, Hollibaugh JT. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aqua. Microb. Ecol. 2003;31:85–98. [Google Scholar]
- 11.Gomez E, Garland JL, Roberts MS. Microbial structural diversity estimated by dilution-extinction of phenotypic traits and T-RFLP analysis along a land-use intensification gradient. FEMS Microbiol. Ecol. 2004;49:253–259. doi: 10.1016/j.femsec.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Wolsing M, Prieme A. Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments. FEMS Microbiol. Ecol. 2004;48:261–271. doi: 10.1016/j.femsec.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann M, Frey B, Kolliker R, Widmer F. Semi-automated genetic analyses of soil microbial communities: comparison of T-RFLP and RISA based on descriptive and discriminative statistical approaches. J. Microbiol. Methods. 2005;61:349–360. doi: 10.1016/j.mimet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Pett-Ridge J, Firestone MK. Redox fluctuation structures microbial communities in a wet tropical soil. Appl. Environ. Microbiol. 2005;71:6998–7007. doi: 10.1128/AEM.71.11.6998-7007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C-P, Ahuja R, Sayler G, Chu K-H. Quantitative molecular assay for fingerprinting microbial communities of wastewater and estrogen-degrading consortia. Appl. Environ. Microbiol. 2005;71:1433–1444. doi: 10.1128/AEM.71.3.1433-1444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan OC, Yang X, Fu Y, Feng Z, Sha L, Casper P, Zou X. 16S rRNA gene analyses of bacterial community structures in the soils of evergreen broad-leaved forests in south-west China. FEMS Microbiol. Ecol. 2006;58:247–259. doi: 10.1111/j.1574-6941.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- 17.Danovaro R, Luna GM, Dell’anno A, Pietrangeli B. Comparison of two fingerprinting techniques, terminal restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis, for determination of bacterial diversity in aquatic environments. Appl. Environ. Microbiol. 2006;72:5982–5989. doi: 10.1128/AEM.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentile ME, Jessup CM, Nyman JL, Criddle CS. Correlation of functional instability and community dynamics in denitrifying dispersed-growth reactors. Appl. Environ. Microbiol. 2007;73:680–690. doi: 10.1128/AEM.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann M, Widmer F. Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices. Appl. Environ. Microbiol. 2006;72:7804–7812. doi: 10.1128/AEM.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hjort K, Lembke A, Speksnijder A, Smalla K, Jansson JK. Community structure of actively growing bacterial populations in plant pathogen suppressive soil. Microb. Ecol. 2007;53:399–413. doi: 10.1007/s00248-006-9120-2. [DOI] [PubMed] [Google Scholar]
- 21.Lazzaro A, Schulin R, Widmer F, Frey B. Changes in lead availability affect bacterial community structure but not basal respiration in a microcosm study with forest soils. Sci. Total Environ. 2006;371:110–124. doi: 10.1016/j.scitotenv.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi Y, Murashima K, Ohara H, Suzuki T, Hayashi H, Sakamoto M, Fukasawa T, Kubota H, Hosono A, et al. Increase in terminal restriction fragments of Bacteroidetes-derived 16S rRNA genes after administration of short-chain fructooligosaccharides. Appl. Environ. Microbiol. 2006;72:6271–6276. doi: 10.1128/AEM.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osborne CA, Rees GN, Bernstein Y, Janssen PH. New threshold and confidence estimates for terminal restriction fragment length polymorphism analysis of complex bacterial communities. Appl. Environ. Microbiol. 2006;72:1270–1278. doi: 10.1128/AEM.72.2.1270-1278.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey J, Ganesan K, Jain RK. Variations in T-RFLP profiles with differing chemistries of fluorescent dyes used for labeling the PCR primers. J. Microbiol. Methods. 2007;68:633–638. doi: 10.1016/j.mimet.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Kvist T, Ahring BK, Westermann P. Archaeal diversity in Icelandic hot springs. FEMS Microbiol. Ecol. 2007;59:71–80. doi: 10.1111/j.1574-6941.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- 26.Siripong S, Rittmann BE. Diversity study of nitrifying bacteria in full-scale municipal wastewater treatment plants. Water Res. 2007;41:1110–1120. doi: 10.1016/j.watres.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 27.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35((Database issue)):D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsh TL, Saxman P, Cole J, Tiedje J. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 2000;66:3616–3620. doi: 10.1128/aem.66.8.3616-3620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricke P, Kolb S, Braker G. Application of a newly developed ARB software-integrated tool for in silico terminal restriction fragment length polymorphism analysis reveals the dominance of a novel pmoA cluster in a forest soil. Appl. Environ. Microbiol. 2005;71:1671–1673. doi: 10.1128/AEM.71.3.1671-1673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarman Simon N. Cleaver: software for identifying taxon specific restriction endonuclease recognition sites. Bioinformatics. 2006;22:2160–2161. doi: 10.1093/bioinformatics/btl330. [DOI] [PubMed] [Google Scholar]
- 31.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33((Database issue)):D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]