Abstract
The development of bioinformatic tools by individual labs results in the abundance of parallel programs for the same task. For example, identification of binding site regions between interacting proteins is done using: ProMate, WHISCY, PPI-Pred, PINUP and others. All servers first identify unique properties of binding sites and then incorporate them into a predictor. Obviously, the resulting prediction would improve if the most suitable parameters from each of those predictors would be incorporated into one server. However, because of the variation in methods and databases, this is currently not feasible. Here, the protein-binding site prediction server is extended into a general protein-binding sites research tool, ProMateus. This web tool, based on ProMate's infrastructure enables the easy exploration and incorporation of new features and databases by the user, providing an evaluation of the benefit of individual features and their combination within a set framework. This transforms the individual research into a community exercise, bringing out the best from all users for optimized predictions. The analysis is demonstrated on a database of protein protein and protein-DNA interactions. This approach is basically different from that used in generating meta-servers. The implications of the open-research approach are discussed. ProMateus is available at http://bip.weizmann.ac.il/promate.
INTRODUCTION
Protein interfaces are drawing much attention of the structural bioinformatics community as well as the rest of the biological world. Many articles have been published classifying complexes according to function, and analyzing the properties that characterize them. Several prediction engines have been developed in order to analyze interfaces, and predict their location for the various interaction types (1–5). The ongoing discussions rising from the literature show large divergence concerning basic aspects. This appears already at the level of how interfaces are defined (change in accessible surface area or various cutoff distances either between heavy atoms or Cα atoms). Further it goes through the definition of successful prediction, which is measured at the level of proteins, amino acids or predefined surface patches (6–11). Finally, it concerns basic issues such as the role of evolutionary conservation in binding that is still controversial (10,12,13). We contributed to this effort, through the development of ProMate (14), a protein-binding sites prediction server. ProMate uses various structural features measured on unbound proteins to identify potential binding sites. Thirteen different properties were examined in ProMate, and using a reduced brute-force optimization, a subset of nine of them was selected to be counted in the final prediction.
At this point, having examined many different alternatives, the field of binding-site prediction has matured to be able to converge to common guiding definitions that are considered most suitable, and are needed in order to focus on the goal itself. A community-wide effort is required for this to be accomplished. Moreover, as research evolves, new insights can improve the prediction. In some cases, the original motivation might not be directly related to the prediction of protein-binding sites rather provide an independent measure related to proteins. In others, the information regarding binding sites location can amplify the significance of a new result. In any case, the value of having a simple tool for testing the relevance of new features to proteins’ binding potential is evident. To this aim, we leveraged ProMate into a generic protein-binding sites analysis web tool, ProMateus. Here, we present this tool and its utilization for three types of results that can be drawn from this type of analysis: improving ProMate's prediction accuracy, extending ProMate for the prediction of protein DNA binding sites, using a relevant training dataset, and for the comparison of a newly suggested definition of secondary structure compositions of proteins based on interaction networks to traditional secondary prediction methods for their binding site occupancy. However, the real goal of ProMateus is to promote a new idea of open research with ProMateus providing an open web tool that facilitates the examination of features that are relevant for binding sites prediction.
RESULTS
ProMateus
Currently, ProMateus allows analyzing new features over three databases: the set of unbound proteins involved in transient-hetero interactions (originally used in ProMate), a bound database of monomeric proteins having a binding site for DNA and a database of models produced for this bound database, to help discarding features that are artifacts of the bound case. Additional, new or altered, databases can be easily integrated. The user, suggesting a new feature that can potentially designate the interface location, should download the relevant database, and upload back the files with the relevant new information (on the whole, or only part of the database). ProMateus will execute a three-phase feature selection procedure to evaluate the contribution of this feature to the prediction success.
Feature selection schemes operate in one of three modes: as filters, as wrappers or as an embedded optimization. Filters are statistical tests that are applied to the data independently of the prediction algorithm. Wrappers are general optimization algorithms that theoretically can be executed with any prediction technique. Embedded algorithms are methods in which the feature selection and the prediction stages are inseparable.
In the first phase, ProMateus uses a simple filtering scheme. A histogram is produced, presenting the distribution of the feature values at interfaces versus the rest of the surface. For categorical features, a bootstrap procedure is used to evaluate the 70% confidence intervals of each category. The histogram of continuous scores is assigned a P-value using the Kolmogorov–Smirnov test. As an alternative, the log file also presents the P-value evaluated from the Pearson's correlation coefficient. If the suggested feature passes this filter namely, the curves are significantly different; ProMateus uses a logistic regression (LR) optimization in 5-fold cross-validation of the new feature together with ProMate's original properties. To simplify the model, the weights assigned by the optimization are limited to the range [0,1], thus no complicated dependencies between features are allowed. Due to the equivalence of ProMate's scoring scheme to LR (explained in the Methods section) this would generally be classified as an embedded scheme. However, note that the LR procedure differs from ProMate by the fact that it acts on the space of the surface dots (see Supplementary Data for detailed description of methods), and not yet at the level of the proteins.
If the suggested feature was assigned a significant weight, in order to reduce noise, a second LR optimization is executed in which features with a weight smaller than one SD from Zero (over the 5-fold split) are eliminated. Finally, these weights are used for the final prediction using ProMate, and the success is measured at the level of the proteins.
The role of the third phase of the optimization is to rule out new features that are overlapping to existing ones. The LR procedure is not biased to prefer as few features as possible; rather, in case of two equivalent features the weight will be divided between them. This by itself can enhance the robustness of the server, since the noise of two different features containing the same information would be reduced. Therefore, the LR optimization is executed as is on all ProMate's existing scores. Since the addition of new features to ProMate would involve additional work, this phase is used in ProMateus to rule out new features that do not contain new information.
In summary, ProMateus returns one of three possible answers: the suggested feature might be irrelevant for interfaces, it might contain information about interfaces, but in a manner that overlaps the features that are already in use, and thus the final prediction is not improved, or it can add orthogonal information that improves the final prediction. In the latter case, the intention is to integrate such scores into ProMate.
Reanalyzing hetero-transient interactions
The original feature optimization used in ProMate was a heavy, brute-force-like optimization procedure, that was limited to choosing features, but did not allow weighting them. In addition to being computationally heavy, such a procedure, risks overfitting to the available database. Using the LR optimization that is limited to a simple model, which is further simplified by limiting the weights to the range of [0,1] downgrades this problem.
The results of re-estimating the features used in ProMate shows some disagreement with its original feature selection. One score is the probability distribution of the different atom types at interfaces. Second is the preference of interfaces to be populated by longer ‘loops’, i.e. unstructured flexible regions of the protein. Also, the sequence distance that is the distance between residues along the peptide chain that tends for the longer distances at interfaces was found significant. Finally, the number of bound water molecules proved to be higher at interfaces already at the unbound structure. All these features are described in detail elsewhere (14). Using these four scores with a weight of 1, the prediction improves from 36 correct predictions out of 51 predictions produced, to 38 out of 55. Thus, an increased coverage was achieved.
In recent years, several other features were claimed to be significant for interface recognition; among those are improved evaluation of evolutionary conservation (such as WHISCY (3) and conSurf (4)), the distance of each atom from the center of mass in enzymes (15), and high-frequency vibrating residues (16). All these features were tested by ProMateus. Specifically, WHISCY was run through its web server, limited to the calculation of the evolutionary score alone, to avoid overlap with the AA propensities which already exist in ProMate. ConSurf was run through its web server, with all the default values except homologs that are collected from UniProt. The distance of each atom from the center of mass was calculated both at the level of atoms, and at the level of amino acids. High-frequency vibrating residues were extracted from the iGNM database (http://ignm.ccbb.pitt.edu/FileDownload.htm) taking the residue mean-square fluctuations driven by the joint contribution of the highest 10 modes. All these features failed before the last phase, namely, though they contain relevant information to interfaces, they do not improve the interface prediction within the suggested model. This conclusion might be inaccurate for the feature of the distance of the atom from the center of mass since the authors claim it should only apply to enzymes, which are a small fraction of the database used. The result of ProMate together with each of these features is presented in Table 1. A predicted interface patch is extracted from the full range of predicted interface probabilities. The bounds for this are optional parameters in ProMateus, allowing the user to experience with a full range of bounds, determining the sensitivity and specificity of the prediction. Throughout our analysis, a prediction is defined successful if it is reliable, namely, if at least 50% of the predicted interface patch is truly so. The number of successful predictions achieved over the proteins is presented in Figure 1. We found a hard core of 20 proteins that were predicted by all the different combinations, 13 proteins that were not predicted by any of the combinations and 24 proteins that were predicted correctly by only some combination. Excluding the distance from the proteins' center of mass, one can see that the scores are overlapping, and the differences are within the noise. Moreover, one clearly sees a difference in the success rate versus coverage. The re-optimized ProMate has the highest coverage (0.96) but a success rate of only 0.69 (resulting in 38 correct predictions), while ProMate + WHISCY has a coverage of only 0.77 but with a success rate of 0.75 resulting in 33 correct predictions. Which of the two choices is better depends whether one needs high coverage or a higher success rate.
Table 1.
Success rate of proMate with new features
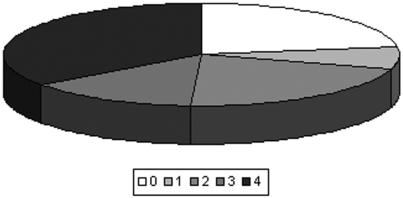
Figure 1.
An overview of the protein–protein interactions database induced by comparing the different methods. The pie chart shows the fraction of proteins where the binding site was predicted by all 4 methods (20 proteins), 3 methods (8), 2 methods (12), 1 method (4) or where all the predictors failed (13 proteins).
Protein–DNA recognition
Protein–DNA-binding sites present an additional class of important biochemical interactions. Their interfaces are similar to protein–protein interfaces in the sense that both span over a large patch on the protein surface, and thus the same nature of analysis would be appropriate. Therefore, we enhanced ProMateus to also apply to them.
The strongest property that characterizes DNA-binding proteins is their positive electrostatic potential used to attract the negatively charged DNA. Studies also incorporated sequential properties such as evolutionary conservation and the frequency of favored residues like lysine and arginine, as well as structural properties of surface curvature and accessible surface area, together with the helix-turn-helix motif that is abundant in DNA-binding proteins (but is not limited to them) (17–20). To demonstrate the great advantage in the simplicity of ProMateus, we utilized the available features used in ProMate and applied them to a database of protein-DNA interfaces. Due to its importance, a simple electrostatic score was added. This potential was constructed by simulating a negative charge at the center of every circle on the protein surface using the program PARE (21) (see Methods section). Since the available unbound database was limited to six proteins, we replaced it by a database of simple models constructed by the fast calculation option of ModWeb, the web server running MODELLER (22). ModWeb assigned each protein from the bound database a template from the PDB, based on sequence similarity. Then, a structure was constructed based on this template and a force-field optimization. As the number of water molecules and temperature factor distribution are biased in case of the bound structures, and unavailable for the models, both were excluded.
The selected features in decreasing importance according to the optimization on the bound database are the distribution of the atoms, the electrostatic potential, the secondary structure, the evolutionary conservation, distribution of AA pairs, single amino acid distribution and finally the chemical character of atoms. A good agreement was observed between the bound and models databases with the main difference being that in the models the importance of the chemical character increased significantly while the weight of the electrostatic potential was low. This is self explanatory by the fact that in order to accurately calculate the electrostatic potential, a high-resolution structure is required. This, of course, would not be the situation in unbound structures, and thus, the models provide a somewhat strict test case, though still of a value by itself.
In the prediction test, only one protein (1qumA) failed to yield any prediction (Table I in the Supplementary Data). For 30 of the remaining 47 proteins the sites were predicted successfully, giving a success rate of 64%. For the models database, 27 out of 46 predictions were correct, i.e. 59% of the database. Going down into the details, 3 proteins (1e7kA, 1ewnA and 1feuA) were predicted successfully for the models, and failed on the bound database. All three models used the original structure as a template for modeling, and the prediction on the bound structure was at the right place, but with a true positive rate lower than 0.5 (0.41, 0.42, and 0.25, respectively). Five proteins succeeded on the bound database, and failed for the models. Three of them were modeled by other proteins with 100% sequence similarity and two of them with lower similarity. For all the five, the binding site was found for the model with a true positive rate of at least 25%. Thus, the agreement between the predictions on the models and bound databases is high.
This article is the first study known to us that deals directly with models. The robustness of ProMateus is exemplified through the fact that the success rate achieved is close to the ones reported, though no specific optimization to DNA-binding proteins was done. The consistency of the feature selection over the two databases validates the method being used.
Secondary structure distributions at interfaces
The role of secondary structures at interfaces has been discussed previously, raising many contradicting conclusions. Jones et al. (23) found α-helices to be favored at interfaces. Gutteridge et al. (24) used it in a neural network aiming to predict interfaces and found it had a low weight in the prediction. In a recent article, Hoskins et al. (25) showed that β-strands that participate in protein–protein interactions exhibit characteristics similar to internal strands rather than regular edge strands, and used this property for interface prediction. In the analysis of ProMate we found interfaces to be richer in β-sheets and poorer in α-helices, in cases where both these structures appear at the same protein. Thus, there is still an uncertainty about the significance that should be associated with the secondary structure in this context.
The classification to secondary structure is an example for a characteristic that could be misleading. Ascribing an amino acid to one of the classes is strongly dependent on its sequence neighbors, and indeed the most popular secondary structure prediction algorithms are based on sequential relations, e.g. Hidden Markov Models. Therefore, we re-analyzed this property more carefully, at the protein level, using hierarchical bootstrapping. The resulting picture is somewhat different (Figure 2). In addition, the sampling at the level of the protein instead of the amino acid widens the confidence intervals, to an uncertainty level. As a counterexample, one can consider the amino acid distribution that does not differ between the two ways of analysis (data not shown).
Figure 2.
Comparison of secondary structure distribution at protein versus amino- acid level. The amino acid secondary structure categories at the upper rows were extracted using PROMOTIF, with H representing helices, G–3–10 helices; E–strands; S–bends; T–turns; B–beta bridges; Small letters stand for edges of the relevant structure. The lower row was extracted using the method by Raveh et al. (26) that is based on contact map clustering. The classification as described by the authors: 1: sheet-like loops; 2: parallel sheets; 3: anti-parallel sheets; 4–6: loops; 7: short and flexible helices; 8: long helices.
In comparison to the traditional definition of secondary structures discussed above, we examine a new definition of secondary structures suggested recently (26). While the common definition used in PROMOTIF (27) is based on predefined angels between consecutive amino acids, the definition suggested by Raveh et al. is based on spatial features extracted by clustering the proteins contact map defined by the backbone and hydrogen bonds. Comparing the distributions based on the two definitions shows that the latter is superior from aspects of the protein function. Class number 4, which is associated with a subset of the loops, shows a significant preference for non-binding surfaces, while no class shows a significant difference with the traditional definition. Thus, the new definition is more relevant from aspects of the protein function.
DISCUSSION
The internet revolution dictates a communal way of research. The simplified communication in the ‘global village’ increases the creation of new knowledge and its utilization around the world. This is in fact one of the driving forces of bioinformatic research. Many servers that provide various scientific services have been established, and are used as daily scientific tools. The vision lying at the base of ProMateus suggests taking this community-research approach one step further.
The industrial community has already acknowledged the advantages of the open source model of system development, in which portions of source-codes are freely distributed by individuals and companies from around the world. In addition to a fast development rate, such projects are considered superior in contribution to world standards, in improved project modularity and even from financial aspects. Inspired by this, ProMateus is an initiative demonstrating the open research approach. The potential of such tools to advance specific areas of research is tremendous, and suggests a way of worldwide research communication that up until now was only available though important contest projects, such as CASP and CAPRI.
The open-research approach differs from the open source by one intrinsic complication that should be acknowledged. When testing many features over the same limited data, the probability of overfitting, i.e. that a random feature would be found significant by chance increases. However, this should not prevent the development of such projects. Employing careful filtering (and cross validation) and updating the available databases should restrain these effects. Taken to the extreme, consider all the structural bioinformatics labs working on one database—the PDB. Applying a careful analysis would enable to gain from a worldwide research effort. Yet, one has to keep in mind the theoretical limitations of such a system, and consider the results carefully.
In the context of binding-site prediction, new features gaining from the expertize of different labs can be easily checked and incorporated into the exiting framework. Another aspect is the creation of standards. One of the hardest problems in the field is the disagreement over the basic definitions, such as those of the binding site itself, and of a successful prediction. Having future similar tools, the inherent natural selection of the web will enable the objective comparison of the various tools and definitions and the convergence to the most promising ones.
The goal of enhancing the understanding of protein recognition is by itself interesting, and important for applications that are not directly related to binding-site prediction. The comparison of the alternative definitions of secondary structures demonstrates this.
This article is the first known to us that analyzes DNA-binding site of protein models. Due to lack of unbound structures, studies in the field usually focus on analyzing the bound structures. Some of them use a small unbound dataset as supporting evidence. Homology models are a neglected class, though these occupy most of the currently available structure space. The goal of predicting the location of the binding site on modeled structures might be more complicated than unbound structures but is certainly worth pursuing. The results in this article showed that the electrostatic potential, considered most important for DNA-binding site identification, significantly loses its strength to a more general chemical character representation when only a model is available. A comparison to a model built on an unbound template is expected to yield further insights.
An important issue that is raised in this article is the alternative ways of analyzing the same property. The analysis at the protein versus AA level exemplifies a problem that most probably rises in many similar studies. Two different sampling models are suggested, and can sometime lead to different conclusions. There is no strict answer regarding which level of analysis is superior. The analysis at the protein level reduces the effect of intra-protein dependencies, but at the same time loses confidence due to smaller sample size. On the other hand, taking the mean over all the proteins removes the bias in favor of larger proteins, but will ascribe higher weight to outlier proteins. When analyzing a new property, one should carefully examine both options, and choose the one which seems more appropriate for the specific case.
To conclude, the main objective of this article is to export the idea of ‘open-research’ into a simple, user-friendly web-based server. Using this idea, the bioinformatics research can leverage above small independent projects and evolve towards fewer centralized worldwide cooperations that integrate different modules contributed by labs around the world. We believe that due to its nature, biological research that often requires high expertize in a small-scale area of research would gain significantly from this approach.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This research, as well as funding to pay the Open Access publication charges for this article were provided by a grant from the Israel Ministry for Science and Technology (MOST) 0263 and by the Sudarsky Center for Computational Biology at the Hebrew University, Jerusalem.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ahmad S, Gromiha MM, Sarai A. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- 2.Bradford JR, Westhead DR. Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics. 2005;21:1487–1494. doi: 10.1093/bioinformatics/bti242. [DOI] [PubMed] [Google Scholar]
- 3.de Vries SJ, van Dijk AD, Bonvin AM. WHISCY: what information does surface conservation yield? Application to data-driven docking. Proteins. 2006;63:479–489. doi: 10.1002/prot.20842. [DOI] [PubMed] [Google Scholar]
- 4.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchiya Y, Kinoshita K, Nakamura H. PreDs: a server for predicting dsDNA-binding site on protein molecular surfaces. Bioinformatics. 2005;21:1721–1723. doi: 10.1093/bioinformatics/bti232. [DOI] [PubMed] [Google Scholar]
- 6.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 7.Bordner AJ, Abagyan R. Statistical analysis and prediction of protein-protein interfaces. Proteins. 2005;60:353–366. doi: 10.1002/prot.20433. [DOI] [PubMed] [Google Scholar]
- 8.Halperin I, Wolfso H, Nussinov R. Protein-protein interactions; coupling of structurally conserved residues and of hot spots across interfaces. Implications for docking. Structure. 2004;12:1027–1038. doi: 10.1016/j.str.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Liang S, Zhang C, Liu S, Zhou Y. Protein binding site prediction using an empirical scoring function. Nucleic Acids Res. 2006;34:3698–3707. doi: 10.1093/nar/gkl454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nooren IM, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang B, Chen P, Huang DS, Li JJ, Lok TM, Lyu MR. Predicting protein interaction sites from residue spatial sequence profile and evolution rate. FEBS Lett. 2006;580:380–384. doi: 10.1016/j.febslet.2005.11.081. [DOI] [PubMed] [Google Scholar]
- 12.Caffrey DR, Somaroo S, Hughes JD, Mintseris J, Huang ES. Are protein-protein interfaces more conserved in sequence than the rest of the protein surface? Protein Sci. 2004;13:190–202. doi: 10.1110/ps.03323604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guharoy M, Chakrabarti P. Conservation and relative importance of residues across protein-protein interfaces. Proc. Natl Acad. Sci. USA. 2005;102:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neuvirth H, Raz R, Schreiber G. ProMate: a structure based prediction program to identify the location of protein-protein binding sites. J. Mol. Biol. 2004;338:181–199. doi: 10.1016/j.jmb.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Shimon A, Eisenstein M. Looking at enzymes from the inside out: the proximity of catalytic residues to the molecular centroid can be used for detection of active sites and enzyme-ligand interfaces. J. Mol. Biol. 2005;351:309–326. doi: 10.1016/j.jmb.2005.06.047. [DOI] [PubMed] [Google Scholar]
- 16.Haliloglu T, Keskin O, Ma B, Nussinov R. How similar are protein folding and protein binding nuclei? Examination of vibrational motions of energy hot spots and conserved residues. Biophys. J. 2005;88:1552–1559. doi: 10.1529/biophysj.104.051342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones S, Thornton JM. Protein–DNA Interactions: the story so far and a new method for prediction. Comp. Funct. Genomics. 2003;4:428–431. doi: 10.1002/cfg.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuchiya Y, Kinoshita K, Nakamura H. Structure-based prediction of DNA-binding sites on proteins using the empirical preference of electrostatic potential and the shape of molecular surfaces. Proteins. 2004;55:885–894. doi: 10.1002/prot.20111. [DOI] [PubMed] [Google Scholar]
- 19.Jones S, Shanahan HP, Berman HM, Thornton JM. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–7198. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad S, Sarai A. In 15th International Conference on Genome Informatics 2004 December 13-15. Japan: Yokohama; 2004. Prediction of DNA-binding sites in proteins using evolutionary profiles. [Google Scholar]
- 21.Selzer T, Schreiber G. Predicting the rate enhancement of protein complex formation from the electrostatic energy of interaction. J. Mol. Biol. 1999;287:409–419. doi: 10.1006/jmbi.1999.2615. [DOI] [PubMed] [Google Scholar]
- 22.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 23.Jones S, Thornton JM. Protein-protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 1995;63:31–65. doi: 10.1016/0079-6107(94)00008-w. [DOI] [PubMed] [Google Scholar]
- 24.Gutteridge A, Bartlett GJ, Thornton JM. Using a neural network and spatial clustering to predict the location of active sites in enzymes. J. Mol. Biol. 2003;330:719–734. doi: 10.1016/s0022-2836(03)00515-1. [DOI] [PubMed] [Google Scholar]
- 25.Hoskins J, Lovell S, Blundell TL. An algorithm for predicting protein-protein interaction sites: abnormally exposed amino acid residues and secondary structure elements. Protein Sci. 2006;15:1017–1029. doi: 10.1110/ps.051589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raveh B, Rahat O, Basri R, Schreiber G. Rediscovering secondary structures as network motifs – an unsupervised learning approach. Bioinformatics. 2007;23:e163–9. doi: 10.1093/bioinformatics/btl290. [DOI] [PubMed] [Google Scholar]
- 27.Hutchinson EG, Thornton JM. PROMOTIF – a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]