Abstract
Bioinformatics testing approaches for protein allergenicity, involving amino acid sequence comparisons, have evolved appreciably over the last several years to increased sophistication and performance. EVALLER, the web server presented in this article is based on our recently published ‘Detection based on Filtered Length-adjusted Allergen Peptides’ (DFLAP) algorithm, which affords in silico determination of potential protein allergenicity of high sensitivity and excellent specificity. To strengthen bioinformatics risk assessment in allergology EVALLER provides a comprehensive outline of its judgment on a query protein's potential allergenicity. Each such textual output incorporates a scoring figure, a confidence numeral of the assignment and information on high- or low-scoring matches to identified allergen-related motifs, including their respective location in accordingly derived allergens. The interface, built on a modified Perl Open Source package, enables dynamic and color-coded graphic representation of key parts of the output. Moreover, pertinent details can be examined in great detail through zoomed views. The server can be accessed at http://bioinformatics.bmc.uu.se/evaller.html.
INTRODUCTION
Allergy, including food allergy, is a major and increasing ailment (1). The disease is strictly associated with atopy, i.e. a genetic predisposition to develop allergic immune reactions to otherwise innocuous components, generally proteins. Several forms of this disorder are described and a major one is designated IgE-mediated allergy, also known as hypersensitivity type I (2). This disease involves reactions to a variety of aerial proteins typically occurring in tree, grass and weed pollen as well as proteins present in a wide range of foods. Animal dander and insect venoms can also cause disease reactions (3). The establishment of allergy consists of two separate phases: sensitation and triggering, i.e. education of the immune system and the actual reaction(s), respectively. The former part involves maturation of naïve T- and B-cells into immunocompetent effector cells, as dictated by a series of complex cellular interactions (4,5). The type-2 helper T-lymphocyte (TH2) has a key function in this process, since it preferentially promotes class switch to IgE-expression. Moreover, a variety of regulatory T-cell subsets play an essential function in the orchestration of an immunological educational procedure (6,7). IgE immunoglobulins can readily bind to high-affinity receptors on tissue mast cells or basophilic granulocytes. The triggering phase is commenced by renewed contact with the antigen, involving binding to cell-anchored IgE molecules and an accordingly elicited release of inflammatory substances, causing anyone or several among a range of symptoms (8–10). Asthma, rhinitis, rhinoconjunctivitis, eczema, contact dermatitis, angioedema and abdominal pain are common allergic reactions, but anaphylactic shock—entailed to impaired respiratory and circulatory function—can also follow.
A sensitized individual may also respond similarly to substances that share certain structural features with the molecule that elicited the initial immune reaction (11–13). This phenomenon, designated cross-reactivity, is tightly connected to the epitopes, i.e. parts of an allergenic protein that are recognized by immunoglobulins—particularly IgE—or receptors present on T-lymphocytes. Broadly defined, such cross-reactivity can engage either IgE- or T-cell epitopes, but that involving IgE-binding (generally referred to as B-cell cross-reactivity) is much better understood (14–16). IgE epitopes can occur either as uninterrupted segments of amino acid residues (continuous epitopes) or distributed as patches on the protein (discontinuous epitopes), the latter sort being brought into juxtaposition in a native (folded) protein configuration. Some common examples of IgE-type cross-reactivity are the pollen-fruit and the latex-fruit syndromes, both categories being associated with promiscuous IgE recognition due to protein structural similarity across species (12,17,18). This phenomenon typically, but not necessarily, occurs between protein allergens from phylogenetically related species (3,19,20). Moreover, a relatively high degree of identity at the amino acid sequence level is commonly seen between IgE cross-reactive proteins (21). Nonetheless, high levels of homology without conservation of allergenicity and low degree of sequence similarity with conservation of the offending property are also reported (20,22).
The complex mechanisms involved in allergy have prompted for several inherently different methods to safely conclude on potential protein allergenicity. Major schemes suggest a tiered set of tests involving amino acid sequence comparison (simple bioinformatics) as well as several in vitro and in vivo assays (23,24). Notably, bioinformatics-type inspection represents a key prescription for allergenicity testing in the subsequently adopted Codex Alimentarius guideline on safety assessment of genetically modified foods and that of the European Food Safety Authority (EFSA) (25,26). The Codex Alimentarius bioinformatics testing scheme, being an early computational design, is built to recognize both general homology-type similarity (to known allergens) and B-cell epitopes; T-cell counterparts may, though, be outside the remit of this allergenicity assessment (25). Intricate relationships between amino acid sequence similarity of query proteins to known allergens and their type-I hypersensitivity potential have, however, spurred further development within this field of risk assessment. Dedicated comparison approaches in conjunction with statistical learning algorithms have enabled increasing overall performance of computational assessment methodology (27–32).
Over the last decade, a variety of Internet-based bioinformatics testing tools for protein potential allergenicity have been developed (31–35). Recently, we reported an in silico method for evaluation of potential protein allergenicity, designated ‘Detection based on Filtered Length-adjusted Allergen Peptides’ (DFLAP) (30). This system is founded on a novel principle, which involves two main features: First, a flexible peptide-selection procedure from allergens, as accomplished by comparing allergens with presumed non-allergens, and secondly, specific education of an support vector machine (SVM) to which a query amino acid sequence can be presented.
In this article, we present EVALLER, a web server-based on a DFLAP core algorithm and an intuitive interface that allows for expedient interrogation of query amino acid sequences, occurring in FASTA format. Subsequent to processing by the DFLAP machine EVALLER returns an assessment of its potential allergenicity, viewed as a rather comprehensive textual and graphical output. EVALLER should be feasible for scanning purposes and as a key part of an integrated allergenicity assessment procedure.
MATERIALS AND METHODS
EVALLER is based on the DFLAP algorithm
The EVALLER server is built as a Perl front end on top of a DFLAP original core, which is implemented in MATLAB. Briefly, the DFLAP algorithm was allowed to construct a set of ‘Filtered Length-adjusted Allergen Peptides’ (FLAPs), derived by extraction from an allergen database through a process involving comparison with a data set (largely compiled from the human proteome) holding proteins with low probability of being allergenic. Selection criteria for both data sets are described elsewhere (30). Amino acid sequences of both allergenic and presumably non-allergenic proteins were subsequently compared with the accordingly extracted FLAPs. Based on the resulting similarity score values for each protein, an SVM was ultimately educated to decide whether a query protein is sufficiently akin to any FLAP to be assigned as an allergen. The allergen data set (762 amino acid sequences), the non-allergen set for FLAP extraction, (52 081 amino acid sequences) and that of presumable non-allergens for DFLAP training (1524 amino acid sequences) were identical to those reported in our earlier study (30). This also applies to DFLAP parameter settings in EVALLER (minimal peptide length lmin = 22, FLAP threshold = 48, number of FLAP matches n = 4 and the cost parameter C = 100 in SVM). For details on these parameters and on the algorithm, see Soeria-Atmadja et al. (30). In its present configuration, EVALLER permits updates of both data and system. Nonetheless, efforts are underway to confer enhanced technical simplicity to this function.
EVALLER assignment uncertainty
A decision statistic, DFLAP score, being an output parameter by SVM algorithm of DFLAP, assigns a query protein as either presumably an allergen or presumably not an allergen depending on whether the aforementioned number is positive or negative. A confidence measure accompanies each risk assessment in order to inform the user on EVALLER's uncertainty regarding assignments. This measure is derived from decision statistics of a test set (not used in the design procedure) holding both allergens and presumed non-allergens. While we needed to hold some data outside the design procedure a DFLAP system was trained using two sets encompassing 500 allergens and 1000 presumed non-allergens, respectively, employed in our earlier published report (30), of which the latter data set was created by random sampling from Swiss-Prot. To achieve minimal incorporation of spurious entries into the non-allergen set, amino acid sequences shorter than 50 amino acid residues or with high sequence similarity to any of the allergens were dismissed. Compliance with the last criterion was accomplished by alignment, allowing a maximum Smith–Waterman score of 100 to be considered as a presumed non-allergen. An identical procedure was applied to compiling a test set of 1000 presumed non-allergens. These test examples are, however, easily assigned as presumable non-allergens by DFLAP. Therefore, additional examples of this category, which are more difficult to assess, were also included (104 vertebrate tropomyosins, 39 animal profilins and 14 mammalian parvalbumins). Of the total 762 allergens the remaining 262 allergens, set apart from the design procedure, were used to estimate decision statistics typical for allergens (all sequence data sets are publicly available on the EVALLER server, under ‘Supplementary Data’). Thus, decision statistics for 1157 (1000+157) presumed non-allergens and 262 allergens were estimated, using the accordingly designed DFLAP system. Uncertainty scores (US) are differently defined for allergen assignments (i.e. the decision statistic DFLAP score >0), relative to those of presumed non-allergens (i.e. the decision statistic DFLAP score <0). Both definitions are specified as the uncertainty, given that the DFLAP score served as a decision threshold instead of zero. In the former (allergen) case, the uncertainty score reflects the probability of false alarms (1-specificity), whereas the probability of overlooking an allergen (1-sensitivity) applies to the latter (non-allergen) situation:
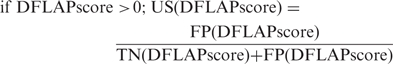 |
1 |
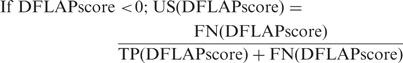 |
2 |
FN(DFLAP score) and TP(DFLAP score) are numbers of misclassified and correctly assigned allergens, respectively, when test statistic DFLAP score is used as a decision threshold. Analogously, FP(DFLAP score) and TN(DFLAP score) are the numbers of incorrect/correct assignments of presumed non-allergens, when DFLAP score applies as a decision threshold.
RESULTS AND DISCUSSION
EVALLER web server
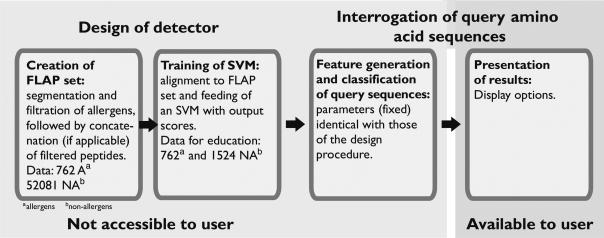
EVALLER was developed to enable bioinformatics assessment of protein potential allergenicity by virtue of the corresponding amino acid sequences. In its present design, protein sequences occurring in FASTA format are accepted. The interrogation procedure, involving submission of a query amino acid sequence (one at a time), allows for user-defined options regarding presentation of results; a range of top-scoring matches and resizable views of the test sequence and matching peptides (Figure 1). Detector design and classification of the test sequence are, however, not available to the user since parameters involved in these procedures have already been optimized (30).
Figure 1.
Flow-chart outlining procedural steps to arrive at the algorithmic core of EVALLER and functions available to the user.
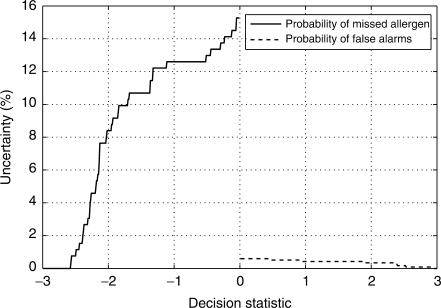
As outlined in ‘Material and Methods’ section, the EVALLER decision statistic for allergenic potential has been split into two separate categories: ‘presumably not an allergen’ and ‘presumably an allergen’. An uncertainty score, indicating the confidence level of EVALLER, is also presented. It is based on the decision statistics from interrogation of a reduced DFLAP, i.e. 262 allergens and 1157 presumed non-allergens (see ‘Material and Methods’ section). An uncertainty score for a negative (presumably a non-allergen) decision statistic, DFLAP score, represents the probability of neglecting an allergen, assuming that the decision threshold had been set at the DFLAP score instead of zero. Analogously, an uncertainty score based on a positive (presumably an allergen) DFLAP score indicates the probability of false alarm, on the assumption that the DFLAP score is identical to the decision threshold. Uncertainty score numbers decrease with increased distance of the DFLAP score to the decision threshold, which is zero (Figure 2). Notably, EVALLER is much more confident in its assignment to the ‘presumably an allergen’ category than that of non-allergens. The uncertainty scores are estimated for a reduced DFLAP (educated with 500 allergens and 1000 presumed non-allergens) rather than for the final DFLAP that EVALLER is based upon (762 allergens and 1524 presumed non-allergens), and may therefore be overly conservative. For clarity, the textual assignment output has been color-coded as determined by category (Figure 3). Green color indicates low allergenic potential whereas red corresponds to a presumable allergen, as judged by EVALLER.
Figure 2.
Graph depicting the level of uncertainty depending on the decision statistic obtained from EVALLER assignment. The uncertainty is defined as the probability of an erroneous assessment assuming the decision threshold is set at the decision statistic instead of zero. Under this condition, the solid line illustrates the probability of neglecting an allergen when EVALLER assigns an amino acid sequence as a presumable non-allergen. The dashed curve illustrates the probability of obtaining a false alarm when EVALLER assigns an amino acid sequence as a presumable allergen.
Figure 3.
Snapshots of the EVALLER output page subsequent to assessment of potential allergenicity for amino acid sequences. Colored (green or red) bars indicate FLAPs, being part of a data set generated by a filtration process as particular for allergenic proteins. The presumably non-allergenic human calmodulin (A) as well as the likely allergenic polcalcin (Syr v 3) from lilac (B). A zoomed-in view focusing between amino residues at position 25 and 50 of the presumed allergen Syr v 3 is also shown (C).
A 2D graphic representation of the scanned sequence, alongside with a color-coded printout (according to the aforementioned assignment scheme) of best matching FLAPs, are presented to the user by virtue of a modified version of the Perl open source package EBioForms (unpublished). The display is dynamic and can be zoomed for detailed views of specific parts of the sequence (Figure 3). Apart from presumed allergen/presumed non-allergen assignment, being integrated in the aforementioned view, EVALLER also provides information on FLAP (see ‘Materials and Methods’ section) to which similarity is identified (Figure 3). Moreover, a link to the UniProt of the cognate allergen accompanies each FLAP. The server and related information is available at the following Internet address: http://bioinformatics.bmc.uu.se/evaller.html.
Benchmarking EVALLER with calcium-binding proteins; comparison with other web servers
The EVALLER core algorithm has been extensively validated for sensitivity and false-alarm rates, using test sets of amino acid sequences (30). Nonetheless, several potential allergens of the polcalcin family of proteins, being members of a widely known protein family involved in pollen–pollen cross-sensitization (36), were not included in those evaluations and may therefore qualify as feasible for a simple test of EVALLER performance. Polcalcins hold two EF-hand calcium-binding domains and typically show high intra-family sequence similarity (37). Presumable non-allergenic homologues to the polcalcin family include the calmodulins and calmodulin-like proteins, as well as other related calcium-binding proteins. An assembly of these proteins, derived from both plant and animal kingdoms, were mined from UniProt and interrogated for potential allergenicity by EVALLER as well as by two additional web servers—Allermatch and APPEL—dedicated to assessment of potential allergenicity (32,34). The former of these already reported methods is designed to apply bioinformatics testing principles outlined by the Codex Alimentarius Commission, i.e. either >35% identity in a segment of 80 amino acid residues (criterion 1) alone or in conjunction with an exact match in stepwise searches for continuous identical sequence segments (criterion 2), whereas the latter implementation derives an assessment from sequence-derived structural and physicochemical properties in combination with an SVM (25,32,34). The EVALLER and APPEL servers assigned all calmodulins or calmodulin-like proteins as presumably non-allergenic, whereas a traditional alignment approach (35% similarity over 80 amino acid segments) gives preference to resemblance of input proteins to known allergens (Table 1). Although EVALLER and Allermatch differ in assignment on (presumably non-allergen) query proteins, both methods identified peptides (or entire amino acid sequences) in allergen polcalcins as best matches. In two cases, however, EVALLER reported differently. The top-scoring FLAP hits for calcium-binding protein-5 (bovine and human) are derived from a troponin-like protein occurring in the fish parasite Anisakis simplex. This protein, however, also contains EF-hand Ca2+ binding motifs, typical of polcalcins, which confers reason to its identification by the EVALLER algorithm (38). Furthermore, to probe for sensitivity of the aforementioned web servers, three polcalcins—neither of them incorporated in the EVALLER system—were submitted for interrogation. Two of them occur in tobacco and one (Syr v 3) in lilac (Table I). The Allermatch database already holds these potential allergens, thereby easily finding perfect matches. Both EVALLER and APPEL, though, assigned Syr v 3 as presumably allergenic, a conclusion supported by a recent report (39). The two tobacco polcalcins were also recognized as potential allergens. For the time being, though, these proteins are devoid of documentation on allergenicity in the scientific literature. Although scored as allergens by EVALLER and the two additional bioinformatics testing tools referred to above, further interrogation is needed to arrive at a scientifically justified conclusion on the allergenicity of the three polcalcin proteins. For this purpose, immunoassay analysis, involving reactivity measurement of IgE antibodies in the sera of patients with clinically validated allergic responses to other polcalcins should apply (25). Other tests, such as resistance to pepsin digestion under appropriate conditions, may further improve assessment accuracy (25,40).
Table I.
Web server benchmarking with calcium binding proteins.
| EVALLER | Allermatcha | APPELb | ||||
|---|---|---|---|---|---|---|
| Query protein | Assignmentc (%) | Best hit | Assignmentd (%) | Best hit | Assignment | Best hit |
| Wheat calmodulin | Not an allergen (13) | Bra n 2 | Allergen (45) | Jun o 4 | Non-allergen | NA |
| Rice calmodulin | Not an allergen (13) | Bra n 2 | Allergen (43) | Jun o 4 | Non-allergen | NA |
| Rape calmodulin | Not an allergen (13) | Bra n 2 | Allergen (46) | Jun o 4 | Non-allergen | NA |
| Mouse calmodulin | Not an allergen (13) | Bra n 2 | Allergen (43) | Jun o 4 | Non-allergen | NA |
| Human CaBP5 | Not an allergen (9) | Ani s Troponin | Allergen (36) | Cyn d 7 | Non-allergen | NA |
| Human calmodulin-like | Not an allergen (13) | Bra n 2 | Allergen (45) | Che a 3 | Non-allergen | NA |
| Human calmodulin | Not an allergen (13) | Bra n 2 | Allergen (43) | Jun o 4 | Non-allergen | NA |
| Bovin CaBP5 | Not an allergen (9) | Ani s Troponin | Allergen (35) | Cyn d 7 | Non-allergen | NA |
| Banana calmodulin-like | Not an allergen (13) | Bra n 2 | Allergen (46) | Jun o 4 | Non-allergen | NA |
| Arabidopsis calmodulin-like | Not an allergen (13) | Bet v 4 | Allergen (44) | Che a 3 | Non-allergen | NA |
| Apple calmodulin | Not an allergen (13) | Bra n 2 | Allergen (45) | Jun o 4 | Non-allergen | NA |
| Tobacco polcalcin 1 | Allergen (0.3) | Ole e 3 | Allergen (100) | Self | Allergen | NA |
| Tobacco polcalcin 2 | Aallergen (0.3) | Ole e 3 | Allergen (100) | Self | Allergen | NA |
| Lilac polcalcin (Syr v 3) | Allergen (0) | Bet v 4 | Allergen (100) | Self | Allergen | NA |
cThe percentage indicates an uncertainty score for the assignment. (See ‘Material and Methods’ section for details.)
dThe percentage indicates the sequence identity of the best 80-mer match, as revealed by alignment (decision threshold is 35%). This is one of the two criteria for bioinformatics assessment of potential allergenicity, as appearing in the Codex Alimentarius guidelines, the other one being scanning for presence of short identical peptide matches.
EVALLER in the context of various approaches to bioinformatics-type risk assessment of protein potential allergenicity
Most of the hitherto accumulated literature on bioinformatics protein allergenicity assessment can be assorted into any of the following several categories: straightforward alignment (e.g. the Codex two-part procedure), alignment-based feature-extraction combined with statistical learning, methods based on comparison to either allergen-derived motifs or reported IgE epitopes and, lastly, similarity search of entire proteins as represented by a variety of coding protocols (27,28,32,41–46). Moreover, by bringing a special kind of segment-reduction procedure into action on allergens, the alignment/statistical learning model has been considerably enhanced (29,30). Hence, EVALLER—built on a DFLAP core—exploits on a tripartite procedure, including a highly specialized filtration of peptides from allergens involving usage of the human proteome, to create material, which permits efficient education of an SVM algorithm. Each output, in response to query submission, is accompanied by an uncertainty numeral, which allows the user to appraise assignment accuracy. Additionally, parts of the query proteins deemed most akin to cognate segments in any or several among the local repository of allergens, are depicted and made available to users of EVALLER in several ways, which altogether support risk assessment.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors wish to thank the Cancer and Allergy Fund, Stockholm, Sweden and the European Model for Bioinformatics Research And Community Education (EMBRACE) network of excellence (grant id: LHSG-CT-2004-512092) for financial support of this work. The European Molecular Biology Network (EMBNET) is acknowledged for providing server facilities. Excellent artwork provided by Ms Merethe Andersen and Hanna Strandberg is greatly appreciated. Funding to pay the Open Access Publication charges for this article was provided by the National Food Administration, Uppsala, Sweden.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hjern A. Chapter 5.8: major public health problems - allergic disorders. Scand J. Public Health Suppl. 2006;67:125–131. doi: 10.1080/14034950600677139. [DOI] [PubMed] [Google Scholar]
- 2.Johansson SG, Bieber T, Dahl R, Friedmann PS, Lanier BQ, Lockey RF, Motala C, Ortega Martell JA, Platts-Mills TA, et al. Revised nomenclature for allergy for global use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003. J. Allergy Clin. Immunol. 2004;113:832–836. doi: 10.1016/j.jaci.2003.12.591. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira F, Hawranek T, Gruber P, Wopfner N, Mari A. Allergic cross-reactivity: from gene to the clinic. Allergy. 2004;59:243–267. doi: 10.1046/j.1398-9995.2003.00407.x. [DOI] [PubMed] [Google Scholar]
- 4.Chaplin DD. 1. Overview of the human immune response. J. Allergy Clin. Immunol. 2006;117:S430–S435. doi: 10.1016/j.jaci.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 5.Romagnani S. Regulatory T cells: which role in the pathogenesis and treatment of allergic disorders? Allergy. 2006;61:3–14. doi: 10.1111/j.1398-9995.2006.01005.x. [DOI] [PubMed] [Google Scholar]
- 6.Aalberse RC, Platts-Mills TA. How do we avoid developing allergy: modifications of the TH2 response from a B-cell perspective. J. Allergy Clin. Immunol. 2004;113:983–986. doi: 10.1016/j.jaci.2004.02.046. [DOI] [PubMed] [Google Scholar]
- 7.Woodfolk JA. T-cell responses to allergens. J. Allergy Clin. Immunol. 2007;119:280–294. doi: 10.1016/j.jaci.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Platts-Mills TA. The role of immunoglobulin E in allergy and asthma. Am. J. Respir. Crit. Care Med. 2001;164:S1–S5. doi: 10.1164/ajrccm.164.supplement_1.2103024. [DOI] [PubMed] [Google Scholar]
- 9.Kay AB. Allergy and allergic diseases. Second of two parts. New Engl. J. Med. 2001;344:109–113. doi: 10.1056/NEJM200101113440206. [DOI] [PubMed] [Google Scholar]
- 10.Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L. The biology of IGE and the basis of allergic disease. Annu. Rev. Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 11.Aalberse RC, Akkerdaas J, van Ree R. Cross-reactivity of IgE antibodies to allergens. Allergy. 2001;56:478–490. doi: 10.1034/j.1398-9995.2001.056006478.x. [DOI] [PubMed] [Google Scholar]
- 12.Yagami T. Allergies to cross-reactive plant proteins. Latex-fruit syndrome is comparable with pollen-food allergy syndrome. Int. Arch. Allergy Immunol. 2002;128:271–279. doi: 10.1159/000063859. [DOI] [PubMed] [Google Scholar]
- 13.Radauer C, Willerroider M, Fuchs H, Hoffmann-Sommergruber K, Thalhamer J, Ferreira F, Scheiner O, Breiteneder H. Cross-reactive and species-specific immunoglobulin E epitopes of plant profilins: an experimental and structure-based analysis. Clin. Exp. Allergy. 2006;36:920–929. doi: 10.1111/j.1365-2222.2006.02521.x. [DOI] [PubMed] [Google Scholar]
- 14.Burastero SE, Paolucci C, Breda D, Longhi R, Silvestri M, Hammer J, Protti MP, Rossi GA. T-cell receptor-mediated cross-allergenicity. Int. Arch. Allergy Immunol. 2004;135:296–305. doi: 10.1159/000082323. [DOI] [PubMed] [Google Scholar]
- 15.Bannon GA, Ogawa T. Evaluation of available IgE-binding epitope data and its utility in bioinformatics. Mol. Nutr. Food Res. 2006;50:638–644. doi: 10.1002/mnfr.200500276. [DOI] [PubMed] [Google Scholar]
- 16.Bohle B. T-cell epitopes of food allergens. Clin. Rev. Allergy Immunol. 2006;30:97–108. doi: 10.1385/CRIAI:30:2:97. [DOI] [PubMed] [Google Scholar]
- 17.Blanco C. Latex-fruit syndrome. Curr. Allergy Asthma. Rep. 2003;3:47–53. doi: 10.1007/s11882-003-0012-y. [DOI] [PubMed] [Google Scholar]
- 18.Egger M, Mutschlechner S, Wopfner N, Gadermaier G, Briza P, Ferreira F. Pollen-food syndromes associated with weed pollinosis: an update from the molecular point of view. Allergy. 2006;61:461–476. doi: 10.1111/j.1398-9995.2006.00994.x. [DOI] [PubMed] [Google Scholar]
- 19.Bowyer P, Fraczek M, Denning DW. Comparative genomics of fungal allergens and epitopes shows widespread distribution of closely related allergen and epitope orthologues. BMC Genomics. 2006;7:251. doi: 10.1186/1471-2164-7-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breiteneder H, Mills C. Structural bioinformatic approaches to understand cross-reactivity. Mol. Nutr. Food Res. 2006;50:628–632. doi: 10.1002/mnfr.200500274. [DOI] [PubMed] [Google Scholar]
- 21.Aalberse RC. Structural biology of allergens. J. Allergy Clin. Immunol. 2000;106:228–238. doi: 10.1067/mai.2000.108434. [DOI] [PubMed] [Google Scholar]
- 22.Mills ENC, Madsen C, Shewry PR, Wichers HJ. Food allergens of plant origin - their molecular and evolutionary relationships. Trends Food Sci. Technol. 2003;14:145–156. [Google Scholar]
- 23.Metcalfe DD, Astwood JD, Townsend R, Sampson HA, Taylor SL, Fuchs RL. Assessment of the allergenic potential of foods derived from genetically engineered crop plants. Crit. Rev. Food Sci. Nutr. 1996;36(Suppl):S165–S186. doi: 10.1080/10408399609527763. [DOI] [PubMed] [Google Scholar]
- 24.FAO/WHO. Rome, Italy: 2001. Evaluation of allergenicity of genetically modified foods. Joint FAO/WHO Expert Consultation on Allergenicity of Foods Derived from Biotechnology. [Google Scholar]
- 25.Codex. Codex Alimentarius. Yokohama: Commission; 2003. Codex Alimentarius Commission. Report of the fourth session of the Codex ad hoc intergovernmental task force on foods derived from biotechnology (ALINORM 03/34A) [Google Scholar]
- 26.EFSA. Guidance document of the Scientific Panel on Genetically Modified Organisms for the risk assessment of genetically modified plants and derived food and feed. EFSA J. 2004;99:1–94. [Google Scholar]
- 27.Zorzet A, Gustafsson M, Hammerling U. Prediction of food protein allergenicity: a bioinformatic learning systems approach. In Silico Biol. 2002;2:525–534. [PubMed] [Google Scholar]
- 28.Soeria-Atmadja D, Zorzet A, Gustafsson MG, Hammerling U. Statistical evaluation of local alignment features predicting allergenicity using supervised classification algorithms. Int. Arch. Allergy Immunol. 2004;133:101–112. doi: 10.1159/000076382. [DOI] [PubMed] [Google Scholar]
- 29.Bjorklund AK, Soeria-Atmadja D, Zorzet A, Hammerling U, Gustafsson MG. Supervised identification of allergen-representative peptides for in silico detection of potentially allergenic proteins. Bioinformatics. 2005;21:39–50. doi: 10.1093/bioinformatics/bth477. [DOI] [PubMed] [Google Scholar]
- 30.Soeria-Atmadja D, Lundell T, Gustafsson MG, Hammerling U. Computational detection of allergenic proteins attains a new level of accuracy with in silico variable-length peptide extraction and machine learning. Nucleic Acids Res. 2006;34:3779–3793. doi: 10.1093/nar/gkl467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha S, Raghava GP. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34:W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui J, Han LY, Li H, Ung CY, Tang ZQ, Zheng CJ, Cao ZW, Chen YZ. Computer prediction of allergen proteins from sequence-derived protein structural and physicochemical properties. Mol. Immunol. 2007;44:514–520. doi: 10.1016/j.molimm.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 33.Ivanciuc O, Schein CH, Braun W. SDAP: database and computational tools for allergenic proteins. Nucleic Acids Res. 2003;31:359–362. doi: 10.1093/nar/gkg010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiers MW, Kleter GA, Nijland H, Peijnenburg AA, Nap JP, van Ham RC. Allermatch, a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines. BMC Bioinformatics. 2004;5:133. doi: 10.1186/1471-2105-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riaz T, Hor HL, Krishnan A, Tang F, Li KB. WebAllergen: a web server for predicting allergenic proteins. Bioinformatics. 2005;21:2570–2571. doi: 10.1093/bioinformatics/bti356. [DOI] [PubMed] [Google Scholar]
- 36.Valenta R, Hayek B, Seiberler S, Bugajska-Schretter A, Niederberger V, Twardosz A, Natter S, Vangelista L, Pastore A, et al. Calcium-binding allergens: from plants to man. Int. Arch. Allergy Immunol. 1998;117:160–166. doi: 10.1159/000024005. [DOI] [PubMed] [Google Scholar]
- 37.Radauer C, Breiteneder H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J. Allergy Clin. Immunol. 2006;117:141–147. doi: 10.1016/j.jaci.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Arrieta I, del Barrio M, Vidarte L, del Pozo V, Pastor C, Gonzalez-Cabrero J, Cardaba B, Rojo M, Minguez A, et al. Molecular cloning and characterization of an IgE-reactive protein from Anisakis simplex: Ani s 1. Mol. Biochem. Parasitol. 2000;107:263–268. doi: 10.1016/s0166-6851(00)00192-4. [DOI] [PubMed] [Google Scholar]
- 39.Ledesma A, Barderas R, Westritschnig K, Quiralte J, Pascual CY, Valenta R, Villalba M, Rodriguez R. A comparative analysis of the cross-reactivity in the polcalcin family including Syr v 3, a new member from lilac pollen. Allergy. 2006;61:477–484. doi: 10.1111/j.1398-9995.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 40.Thomas K, Aalbers M, Bannon GA, Bartels M, Dearman RJ, Esdaile DJ, Fu TJ, Glatt CM, Hadfield N, et al. A multi-laboratory evaluation of a common in vitro pepsin digestion assay protocol used in assessing the safety of novel proteins. Regul. Toxicol. Pharmacol. 2004;39:87–98. doi: 10.1016/j.yrtph.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Hileman RE, Silvanovich A, Goodman RE, Rice EA, Holleschak G, Astwood JD, Hefle SL. Bioinformatic methods for allergenicity assessment using a comprehensive allergen database. Int. Arch. Allergy Immunol. 2002;128:280–291. doi: 10.1159/000063861. [DOI] [PubMed] [Google Scholar]
- 42.Kleter GA, Peijnenburg AA. Screening of transgenic proteins expressed in transgenic food crops for the presence of short amino acid sequences identical to potential, IgE - binding linear epitopes of allergens. BMC Struct. Biol. 2002;2:8. doi: 10.1186/1472-6807-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadler MB, Stadler BM. Allergenicity prediction by protein sequence. FASEB J. 2003;17:1141–1143. doi: 10.1096/fj.02-1052fje. [DOI] [PubMed] [Google Scholar]
- 44.Li KB, Issac P, Krishnan A. Predicting allergenic proteins using wavelet transform. Bioinformatics. 2004;20:2572–2578. doi: 10.1093/bioinformatics/bth286. [DOI] [PubMed] [Google Scholar]
- 45.Ivanciuc O, Schein CH, Braun W. Data mining of sequences and 3D structures of allergenic proteins. Bioinformatics. 2002;18:1358–1364. doi: 10.1093/bioinformatics/18.10.1358. [DOI] [PubMed] [Google Scholar]
- 46.Schein CH, Ivanciuc O, Braun W. Common physical-chemical properties correlate with similar structure of the IgE epitopes of peanut allergens. J. Agric. Food Chem. 2005;53:8752–8759. doi: 10.1021/jf051148a. [DOI] [PubMed] [Google Scholar]