Abstract
The side chains of the 20 types of amino acids, owing to a large extent to their different physical properties, have characteristic distributions in interior/surface regions of individual proteins and in interface/non-interface portions of protein surfaces that bind proteins or nucleic acids. These distributions have important structural and functional implications. We have developed accurate methods for predicting the solvent accessibility of amino acids from a protein sequence and for predicting interface residues from the structure of a protein-binding or DNA-binding protein. The methods are called WESA, cons-PPISP and DISPLAR, respectively. The web servers of these methods are now available at http://pipe.scs.fsu.edu. To illustrate the utility of these web servers, cons-PPISP and DISPLAR predictions are used to construct a structural model for a multicomponent protein–DNA complex.
INTRODUCTION
The growth of protein structures in the Protein Data Bank (PDB) and expansion of our understanding of protein physical properties are constantly enhancing our ability to predict structural and functional features. Many prediction methods are now automated and accurate. We have contributed three such methods: WESA for predicting the solvent accessibility of amino acids from a protein sequence (1), cons-PPISP for predicting interface residues from the structure of a protein which binds a second protein (2,3), and DISPLAR for predicting interface residues from the structure of a protein which binds DNA (4). Both WESA and DISPLAR were found to have higher prediction accuracy than competing methods (1,4). Cons-PPISP was shown to be able to complement experimental techniques such as NMR chemical shift perturbation in mapping protein–protein interfaces (3). The methods have found uses in protein structure prediction (5) and in docking of protein complexes (6). For wide access to them, we have now developed web servers for these methods. Here we describe the functionality of the web servers and illustrate their utility by a structural model, built from predictions of the web servers, for a multicomponent protein–DNA complex.
The web servers are components of PI2PE, the protein interface/interior prediction engine, located at http://pipe.scs.fsu.edu. Together, they serve as a pipeline from protein sequences to tertiary structures, then onto quaternary structures of binary complexes, and finally onto superstructures of functioning, multicomponent complexes.
The three prediction methods have similar designs. The input in each case consists of data for a list of residues. In WESA, the list is comprised of a central residue and equal numbers of its ‘sequential’ neighbors on the left and on the right. In both cons-PPISP and DISPLAR, the list is comprised of a central residue and a number of its ‘spatial’ neighbors. The input data in WESA are sequential profiles, as given by the position-specific scoring matrix produced by PSI-blast (7). In cons-PPISP and DISPLAR, solvent accessibilities (defined as percentages of exposed surface areas of residues) are also included in the input.
METHODS
All the three prediction methods used structures collected from the PDB for training. The dataset of WESA was comprised of 2148 proteins chains with sequence identity <25%. For cons-PPISP, the dataset was comprised of 1256 protein chains that form either heterodimers (involving 458 chains) or homodimers (accounting for the remaining 798 chains of the dataset). The sequence identities among the 1256 chains were <30%. For DISPLAR, the dataset was comprised of 264 protein multimers that form complexes with DNA in their PDB entries. The identities among multimeric entries were <50%. The three protein lists can be downloaded at http://pipe.scs.fsu.edu.
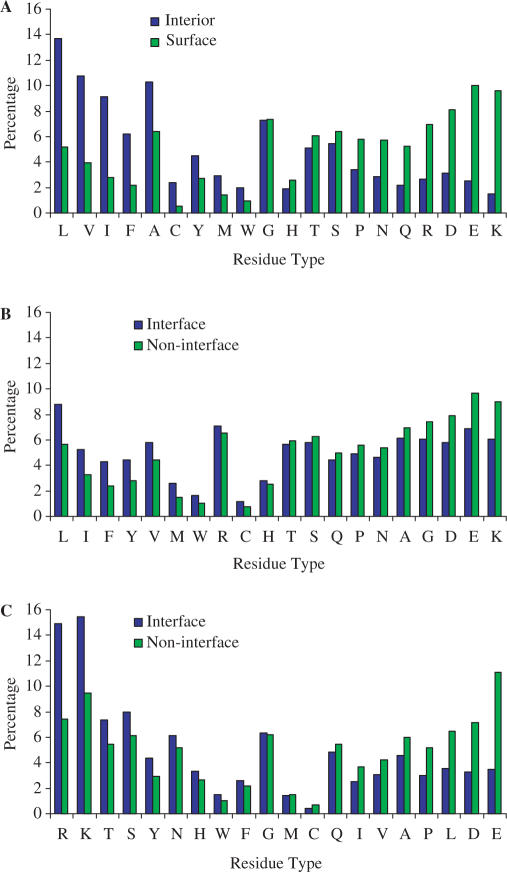
To illustrate why the prediction methods work, in Figure 1 we show the distributions of the 20 types of amino acids in interior and surface regions of folded proteins, and in interface and non-interface portions of protein surfaces that bind proteins or DNA. The results were calculated on the respective datasets. It is clear that the distributions exhibit distinctive patterns. Expectedly non-polar amino acids show preference for the interior whereas polar and charged residues for the surface. Non-polar amino acids similarly (albeit less prominently) prefer interfaces of protein–protein complexes. On the other hand, protein–DNA interfaces appear to be more dictated by electrostatic complementarity (instead of hydrophobicity), with positively charged arginine and lysine significantly enriched while negatively charged aspartate and glutamate significantly depleted in the interfaces.
Figure 1.
Percentages of the 20 types of amino acids in (A) interior and surface regions of folded proteins, (B) interface and non-interface portions of protein surfaces in protein–protein complexes, and (C) interface and non-interface portions of protein surfaces in protein–DNA complexes. In each plot, the amino acids are ordered according to the difference between the two contrasting groups. Interior (or surface) residues were those with < (or >) 20% solvent accessibilities; interface residues were those with a heavy atom that is <5 Å from a heavy atom across a protein–protein or protein–DNA interface.
Another feature, captured by sequence profiles, that distinguishes interior from surface and interface from non-interface is sequence conservation. For structural and functional reasons, interior and interface positions are expected to be more conserved. Such a trend is indeed shown through a comparison in conservation scores between interior and surface positions and between interface and non-interface positions (Supplementary Figure 1).
In addition to sequence profiles, cons-PPISP and DISPLAR also use solvent accessibilities as part of the input. In protein–protein complexes, interface positions consistently have higher solvent accessibilities than non-interface positions. On the other hand, in protein–DNA complexes, positively charged arginine and lysine have higher solvent accessibilities in interface positions than in non-interface positions, but negatively charged aspartate and glutamate show the opposite tendency (Supplementary Figure 2).
Both cons-PPISP and DISPLAR are based on training neural networks. WESA is a metamethod, based on a weighted ensemble of five separate methods, one of which is neural network training. Further details on the implementations of the methods can be found in the original papers (1–4). WESA has consistently shown a two-state (interior/surface) prediction accuracy ∼80%. Cons-PPISP predictions cover >50% of actual protein–protein interface residues and have >70% accuracy. DISPLAR predictions cover >60% of actual protein–DNA interface residues and have >80% accuracy.
FUNCTIONALITY OF THE WEB SERVERS
The direct web link for the WESA web server is http://pipe.scs.fsu.edu/wesa.html. Once there, the user is asked to provide the sequence, in FASTA format, of the protein on which solvent accessibility is predicted. In addition, the user is asked to type in a unique identifier, at the user's choice, for referencing the particular WESA submission, and an e-mail address, for receiving the prediction. A link (http://pipe.scs.fsu.edu/output_wesa.txt) also provides a sample output (obtained from submitting the sequence of PDB entry 1who), with explanations for the columns of numbers for each residue. In short, for each residue in the sequence, the results predicted by five separate methods and the WESA metamethod are given as 1 for exposed or 0 for buried, along with the prediction confidence (ranging from 0.00 for no confidence at all to 1.00 for full confidence). The threshold for an exposed residue is set at 20% solvent exposure. Two figures displaying the actual and WESA-predicted buried residues of 1who are found at the web server.
The direct web link for the cons-PPISP web server is http://pipe.scs.fsu.edu/ppisp.html. The protein structure, on which an interface prediction is to be made, must be provided in PDB format, either by uploading or by pasting. In addition, the user must specify the chain(s) in the structure to be used for prediction. Here there are three common possibilities. (1) The PDB file does not have chain ID; “_”is to be entered. (2) The PDB file has a single chain ID (say “A”), or, it has multiple chains but only a single chain is to be used for prediction; “A” must be entered. (3) Multiple chains (e.g. A, B and C) in a PDB file are to be treated as a single structure and used together for interface prediction; the user must enter “A,B,C”. A sample output can be found at http://pipe.scs.fsu.edu/output_ppisp.txt.
The direct web link for the DISPLAR web server is http://pipe.scs.fsu.edu/displar.html. Input and output formats are very similar to cons-PPISP predictions.
For convenience of using the web servers, we provide scripts for running predictions in a batch mode. The scripts allow the user to submit multiple jobs by a unix command from his/her terminal, as if the prediction programs are installed on the local computer. In reality the scripts upload the sequences or PDB files to the PI2PE web servers. The predictions are run at the servers, and results are sent back to the local computer and saved in files specified by the scripts.
ILLUSTRATIVE APPLICATIONS
We now present interface residues predicted by the cons-PPISP and DISPLAR web servers to illustrate their utility. First, results are given for three proteins that competitively bind proteins and DNA and have unbound structures in the PDB. Next, predictions for a protein that simultaneously binds DNA and another protein are given, and the results are used to build structures for the protein–protein binary complex and the protein–DNA ternary complex. Full lists of predicted interface residues for the proteins are given in Supplementary Table S1.
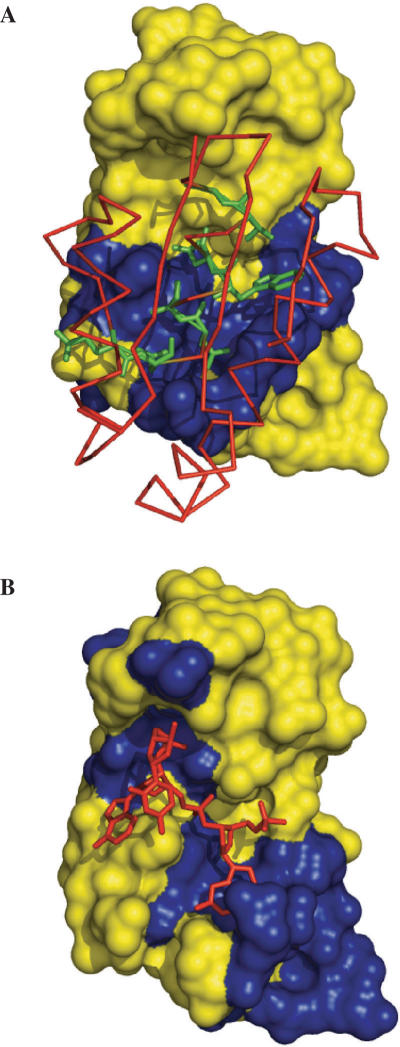
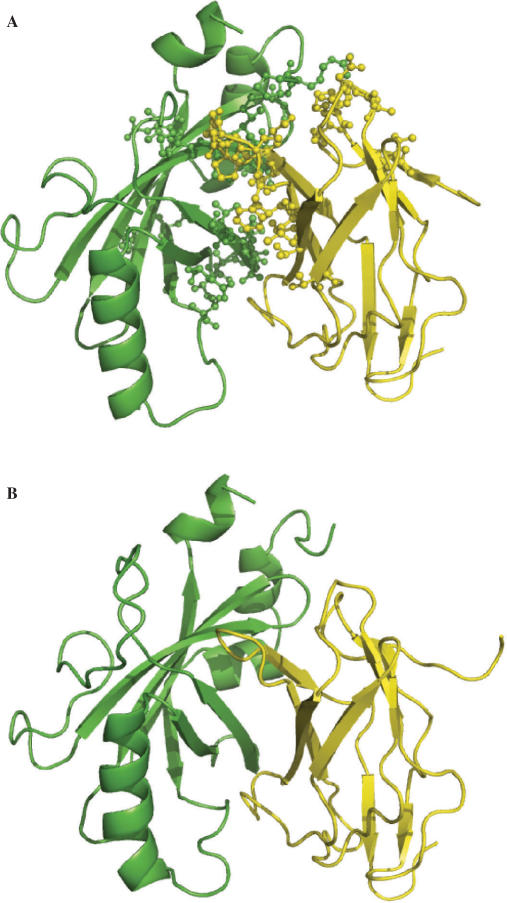
Figure 2A displays the cons-PPISP predictions for the ribonuclease barnase, in reference to the structure of the complex with its inhibitor, barstar (PDB entry 1brs). The predictions were made on the unbound structure of barnase, 1a2p. Twenty predicted residues line up the actual interface. DISPLAR also correctly predicted the same portion of the barnase surface for binding DNA. As shown in Figure 2B, the 27 predicted interface residues are concentrated around the binding site for a tetradeoxynucleotide, d(CGAC) (as defined in 1brn). Six residues (I55-S57, R59, Y97 and Y103) are common in the two sets of predictions. It is of interest to note that cons-PPISP predicted two clusters on the unbound structure of barstar (1bta), one is in the actual interface with barnase (shown in Figure 2A) and the other defines an unknown binding site. DISPLAR did not make any positive predictions on 1bta, which is not expected to bind DNA.
Figure 2.
Predictions of residues in (A) barnase–barstar and (B) barnase–DNA interfaces by cons-PPISP and DISPLAR, respectively, shown on the X-ray structures of the complexes (1brs and 1brn). Predictions were made on unbound structures (1a2p for barnase and 1bta for barstar). Barnase is shown in surface, with predicted interface residues in blue and the rest of the surface in yellow. Barstar and d(CGAC) are shown as red sticks; side chains of barstar residues predicted in the interface with barnase are shown as green sticks. This and Figures 3–6 are generated with PyMOL (http://www.pymol.org).
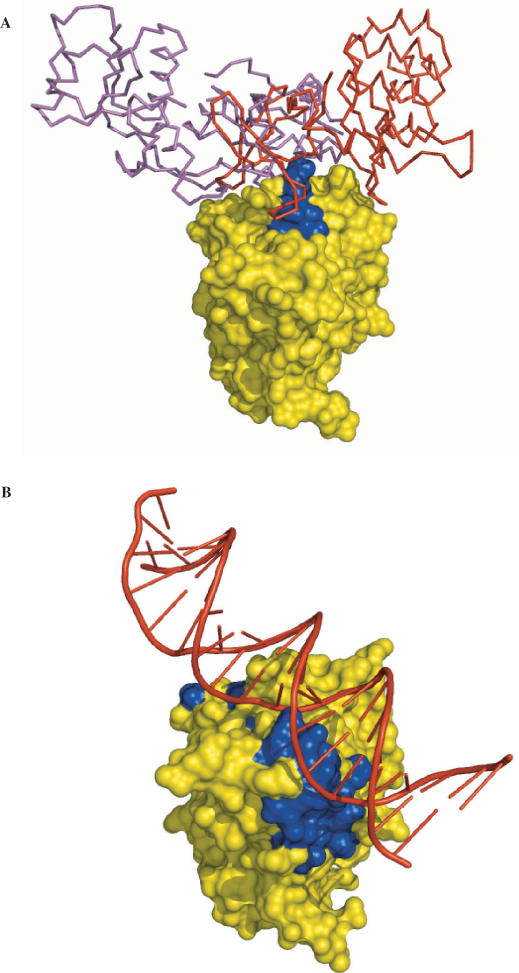
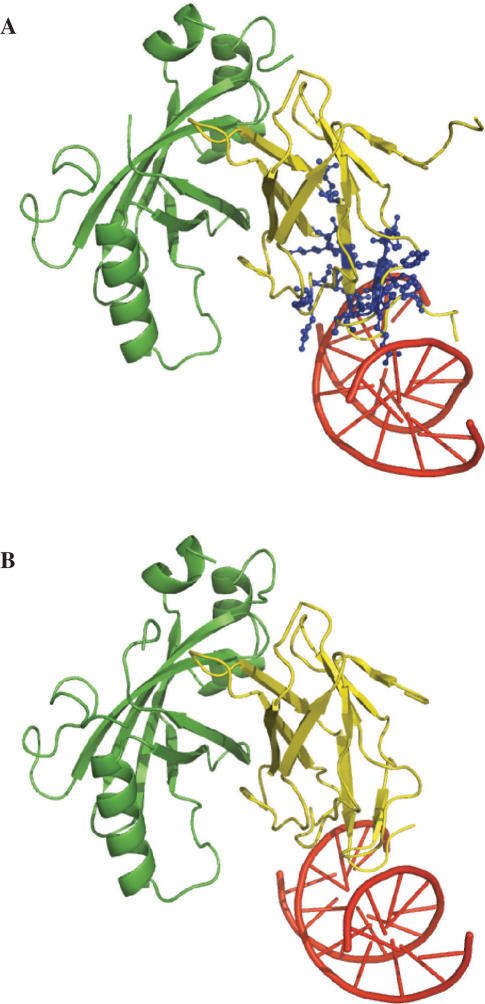
We applied cons-PPISP to the tumor repressor p53 core domain (2fej). In Figure 3A, the predictions are displayed on the structure of p53 in complex with either 53BP1 or 53BP2 (1gzh or 1ycs). The five predicted residues (N239-M243) lie in the interfaces with the p53-binding proteins. A second cluster of 22 residues (listed in Supplementary Table 1) was also predicted; many of these residues are found in the dimer–dimer interface of a tetramer of the p53 core domain bound to DNA (8). Some of these residues are also implicated in the binding with the E6/E6-AP complex (9). When DISPLAR was applied on 2fej, 19 residues were predicted. These also lie in the interface with DNA (as found in 1tsr) (Figure 3B). The protein- and DNA-binding sites on p53 partly overlap. Cons-PPISP predicted residues are close to some of the DISPLAR predicted residues, but no residue was predicted by both methods.
Figure 3.
Protein-contacting and DNA-contacting residues predicted by cons-PPISP and DISPLAR, respectively, on the p53 core domain. Predictions were made on the unbound structure of p53 (2fej) but are displayed on the bound (A) protein–protein and (B) protein–DNA (1tsr) complexes. p53 is shown as surface, with predicted residues in blue and the rest of the surface in yellow. In (A) the structures of the complexes of p53 with 53BP1 (1gzh) and 53BP2 (1ycs), after superimposing p53, are shown, with 53BP1 and 53BP2 as purple and red sticks, respectively.
The two-component transcriptional activator PhoB regulates its DNA-binding activity through transiently forming a domain–domain complex, thereby blocking the DNA-binding site. We used cons-PPISP to predict interface residues on the unbound structures of both the effector and receiver domains (1qqi and 1b00). The structure for full-length PhoB is not available, but we found the structure (1ys6) for a close homolog, PrrA, in the PDB. The root-mean-square deviations (RMSD) of 1qqi and 1b00 from the corresponding domains in 1ys6 are 2.4 and 2.1 Å, with sequence identities over aligned positions at 35 and 41%, respectively. When 1qqi and 1b00 are aligned onto 1ys6, the 23 and 21 predicted residues on the two respective domains indeed mostly line the interface as modeled on 1ys6 (Figure 4A). DISPLAR predicted 25 residues on 1qqi, which are located in the actual interface between the effector domain and DNA (as found in 1gxp) (Figure 4B). The binding sites for the receiver domain and DNA on the effector domain (1qqi) overlap, and eight residues (R68, T69, D71, H73 and V93-T96) were predicted by both cons-PPISP and DISPLAR. No DNA-contacting residues were predicted by DISPLAR on the receiver domain (1b00), which is not known to binding DNA.
Figure 4.
(A) Predicted protein-contacting residues on the effector and receiver domains of PhoB. The two domains are shown as yellow and green ribbons, respectively; side chains of predicted residues are shown as ball-and-stick. The two unbound structures (1qqi and 1b00) are superimposed to the respective domains in PrrA (1ys6). (B) DNA-contacting residues predicted on the unbound structure of the PhoB effector domain (1qqi) and displayed on the complex with DNA (1gxp).
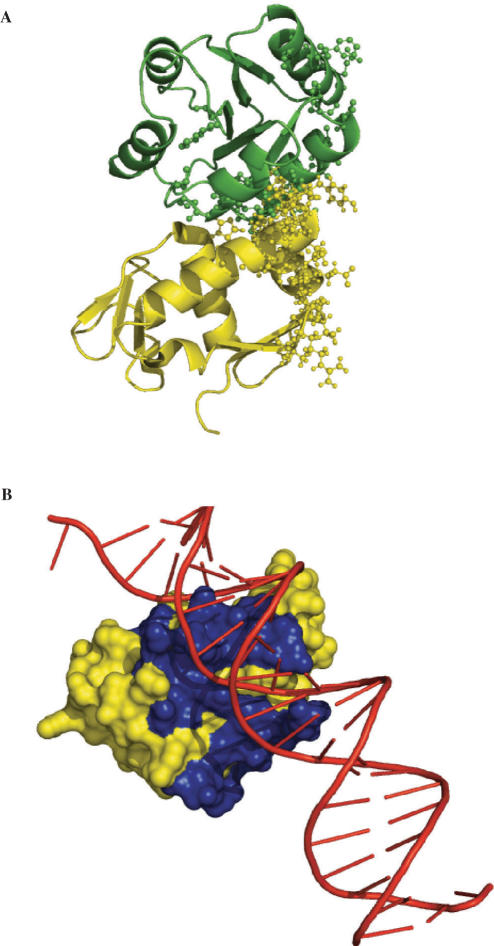
Core binding factors (CBF) form a heterodimer between the α and β subunits, which in turn forms a ternary complex with DNA. Using the unbound structures of CBFα and CBFβ (1eaq and 1ilf, respectively), we predicted interface residues between these two proteins by cons-PPISP. Examined on the dimer structure of CBFα and CBFβ (1e50), the 20 and 23 predicted residues on the two subunits indeed line the actual interface (Figure 5A). Previously we have used cons-PPISP predictions to drive the docking of unbound structures (6). Here we use the predictions to score docked structures, obtained by running ZDOCK 2.3 (10) with 15° sampling. The best 200 ZDOCK structures (out of a total of 2000) were re-ranked according to the number of cons-PPISP predicted residues among the interfacial residues (defined as having 10-Å contacts across the interface). A structure with an RMSD of 2.2 Å was ranked second according to cons-PPISP predictions (improved from ZDOCK's ranking of 49th). This docked structure is shown in Figure 5B.
Figure 5.
(A) Protein-contacting residues on CBFα and CBFβ (as yellow and green ribbons, respectively), predicted on the unbound structures (1eaq and 1ilf) and displayed on the dimeric complex (1e50) as ball-and-stick. (B) Structure for the complex obtained by docking 1eaq and 1ilf and re-ranking according to predicted interface residues.
The dimer structure of CBFα and CBFβ, 1e50, was used to predict DNA-contacting residues by DISPLAR. Nineteen residues, all on CBFα, were predicted. As shown in Figure 6A, CBFα is indeed the subunit that contacts DNA in the ternary complex (1h9d), and the predicted CBFα residues line the DNA-binding site. We docked the dimer structure with a B-DNA decamer built in InsightII (Accelrys Software Inc., San Diego, CA, USA) from the sequence in 1h9d. Parameters for DNA nucleotides required for running ZDOCK were taken from Fanelli and Ferrari (11). A docked structure for the ternary complex, ranked 113th in ZDOCK but improved to 11th by DISPLAR predictions, was found to have an RMSD of just 1.2 Å from 1h9d and is shown in Figure 6B.
Figure 6.
(A) DNA-contacting residues predicted on the dimeric complex of CBFα and CBFβ (1e50), and displayed on the ternary complex (1h9d). Side chains of predicted residues, exclusively located on CBFα, are shown as blue ball-and-stick. (B) Structure for the ternary complex obtained by docking 1e50 and a modeled B-DNA decamer and re-ranking according to predicted interface residues.
FUTURE DEVELOPMENTS
The continuous growth of the sequence database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) and the PDB (http://www.rcsb.org) will further improve the accuracy of the three prediction methods. We plan to periodically upload the NCBI the non-redundant (nr) onto PI2PE (http://pipe.scs.fsu.edu). In addition, we plan to expand the datasets for the three predictors by including new entries from the PDB; re-training will be done.
The high accuracy of WESA predictions suggests that solvent accessibility is now a matured field. Future methodological developments will thus focus on protein–protein and protein–DNA interface predictions. A strategy that contributed to the success of WESA is the combination of complementary methods. We plan to combine cons-PPISP and DISPLAR with approaches based on phylogenetic tree (12,13), surface patch characteristics (14), secondary structure (15), empirical scoring function (16), support vector machine (17–20) and Bayesian network (21,22).
Ultimately proteins need to be studied within their functioning units, which often are multicomponent protein complexes. The PI2PE web servers will contribute to better understanding of these complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Jeff McDonald for assistance in setting up the web servers. This work was supported in part by NIH grant GM058187. Funding to pay the Open Access publication charges for this article was provided by the NIH (grant GM058187).
Conflict of interest statement. None declared.
REFERENCES
- 1.Chen H, Zhou H-X. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33:3193–3199. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou H-X, Shan Y. Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins. 2001;44:336–343. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Zhou H-X. Prediction of interface residues in protein–protein complexes by a consensus neural network method: Test against NMR data. Proteins. 2005;61:21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- 4.Tjong H, Zhou H-X. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007;35:1465–1477. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007 doi: 10.1002/prot.21702. (in press) [DOI] [PubMed] [Google Scholar]
- 6.van Dijk ADJ, de Vries SJ, Dominguez C, Chen H, Zhou H-X, Bonvin AMJJ. Data-driven docking: HADDOCK's adventures in CAPRI. Proteins. 2005;60:232–238. doi: 10.1002/prot.20563. [DOI] [PubMed] [Google Scholar]
- 7.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Wang PL, Sait F, Winter G. The ‘wildtype’ conformation of p53: epitope mapping using hybrid proteins. Oncogene. 2001;20:2318–2324. doi: 10.1038/sj.onc.1204316. [DOI] [PubMed] [Google Scholar]
- 10.Chen R, Li L, Weng Z. ZDOCK: an Initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 11.Fanelli F, Ferrari S. Prediction of MEF2A–DNA interface by rigid body docking: a tool for fast estimation of protein mutational effects on DNA binding. J. Struct. Biol. 2006;153:278–283. doi: 10.1016/j.jsb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan DH, Kristensen DM, Mittleman D, Lichtarge O. ET viewer: an application for predicting and visualizing functional sites in protein structures. Bioinformatics. 2006;22:2049–2050. doi: 10.1093/bioinformatics/btl285. [DOI] [PubMed] [Google Scholar]
- 14.Murakami Y, Jones S. SHARP2: protein–protein interaction predictions using patch analysis. Bioinformatics. 2006;22:1794–1795. doi: 10.1093/bioinformatics/btl171. [DOI] [PubMed] [Google Scholar]
- 15.Hoskins J, Lovell S, Blundell TM. An algorithm for predicting protein–protein interaction sites: abnormally exposed amino acid residues and secondary structure elements. Protein Sci. 2006;15:1017–1029. doi: 10.1110/ps.051589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang S, Zhang C, Liu S, Zhou Y. Protein binding site prediction using an empirical scoring function. Nucleic Acids Res. 2006;34:3698–3707. doi: 10.1093/nar/gkl454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford JR, Westhead DR. Improved prediction of protein–protein binding sites using a support vector machines approach. Bioinformatics. 2005;21:1487–1494. doi: 10.1093/bioinformatics/bti242. [DOI] [PubMed] [Google Scholar]
- 18.Bordner AJ, Abagyan R. Statistical analysis and prediction of protein–protein interfaces. Proteins. 2005;60:353–366. doi: 10.1002/prot.20433. [DOI] [PubMed] [Google Scholar]
- 19.Kuznetsov IB, Gou Z, Li R, Hwang S. Using evolutionary and structural information to predict DNA-binding sites on DNA-binding proteins. Proteins. 2006;64:19–27. doi: 10.1002/prot.20977. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford JR, Needham CJ, Bulpitt AJ, Westhead DR. Insights into protein–protein interfaces using a Bayesian network prediction method. J. Mol. Biol. 2006;362:365–386. doi: 10.1016/j.jmb.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 22.Yan C, Terribilini M, Wu F, Jernigan RL, Dobbs D, Honavar V. Predicting DNA-binding sites of proteins from amino acid sequence. BMC Bioinformatics. 2006;7:262. doi: 10.1186/1471-2105-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]