Abstract
Here we present firestar, an expert system for predicting ligand-binding residues in protein structures. The server provides a method for extrapolating from the large inventory of functionally important residues organized in the FireDB database and adds information about the local conservation of potential-binding residues. The interface allows users to make queries by protein sequence or structure. The user can access pairwise and multiple alignments with structures that have relevant functionally important binding sites. The results are presented in a series of easy to read displays that allow users to compare binding residue conservation across homologous proteins. The binding site residues can also be viewed with molecular visualization tools. One feature of firestar is that it can be used to evaluate the biological relevance of small molecule ligands present in PDB structures. With the server it is easy to discern whether small molecule binding is conserved in homologous structures. We found this facility particularly useful during the recent assessment of CASP7 function prediction. Availability: http://firedb.bioinfo.cnio.es/Php/FireStar.php.
INTRODUCTION
Genome sequencing projects have lead to a surge in the number of protein sequences that lack experimental functional data. At the same time the rise of structural genomics initiatives has meant that the structural databases are starting to fill up with unannotated structures. Experimental approaches for function characterization are expensive and difficult to automate and this has meant that researchers have turned increasingly to computational methods to try to close the gap between the number of new unannotated sequences and the number of sequences with known function.
The standard way to overcome this deficit is the homology-based transfer of functional annotation. The transfer of general functional information [in the form of GO terms (1), etc.] typically requires little more than a BLAST (2) homology search, as long as the protein does not have more than one domain and as long as the query sequence meets a certain threshold of similarity to the annotated protein template. The main constraint for this classical approach is that transfer of function based solely on the percentage of identity of two sequences is not always 100% reliable (3). This constraint has meant that it has been necessary to develop techniques that are capable of assigning function in a more sophisticated manner.
In many cases the most interesting functional information, such as catalytic residues and those residues bound to ligands, is to be found at the residue level. Here transference of function is considerably more laborious since binding sites are disperse, and alignments must be generated and checked by hand before the interesting residues can be mapped onto the query sequence.
Homology-based transfer of functional information at the residue level has been explored in numerous works. The Catalytic Site Atlas (4) is built from annotations extracted from literature and these annotations are extended to the whole PDB trough PSI-BLAST transference. The same authors used this to explore the evolution of catalytic sites in homologous families (5). Automated functional information transfer is also carried out by databases such as Swissprot-Trembl (6). However, while there are web servers that use homology to predict functional features such as GO terms (7–9), and web servers that predict probable binding sites based on clefts or cavities (10) we do not know of an available server that is capable of mapping functional residues onto a target with the aim of highlighting potential functionally important residues.
Here we present firestar, an expert system that merges the time consuming tasks of alignment and mapping into a single server with a simple input. It combines the FireDB (11) database, a large inventory of structure-based functionally important residues, and SQUARE (12,13), a method for the assessment of the local reliability in sequence alignments, to predict likely residues of functional importance in query sequences. This simple tool also includes measures of reliability for the predictions, a set of methods for the visualization of the results on the corresponding sequences and structures and a multiple alignment option for easy comparison.
METHODS
FireDB
The FireDB database is a databank containing a comprehensive and detailed repository of known functionally important residues. It integrates biologically relevant data filtered from the close atomic contacts in Protein Data Bank (14) crystal structures and reliably annotated catalytic residues from the Catalytic Site Atlas. Residues in close contact with ligands are defined as those residues with atoms that are closer than 1.0Å plus the sum of Van der Waals radius of the atoms involved.
Redundancy in the PDB was addressed when designing the database. PDB sequences are clustered with cd-hit (15) at 97% sequence identity and a consensus sequence is built for each cluster. The consensus sequences form the basis of FireDB and of firestar. All functional information is associated to the consensus sequences—equivalent binding sites from proteins within the same cluster are conveniently mapped onto the consensus sequence. The functional residues used by firestar are derived from all the proteins in each cluster, but associated to just a single sequence. As of 8 January 2007, FireDB contained a total of 16 843 clusters, of which 9021 had associated functional information.
SQUARE
firestar evaluates the probability that a residue is involved in ligand binding with a version of SQUARE, a method that was developed to predict regions of reliably aligned residues in pairwise sequence alignments. For SQUARE to evaluate the reliability of an alignment one of the two sequences in the alignment must be associated with a PSI-BLAST-generated profile. In the case of firestar, PSI-BLAST profiles are pre-generated for all the FireDB consensus sequences. This allows SQUARE to evaluate all the pairwise alignments in firestar—the sequence and structural alignments generated as part of firestar are all between the query sequence and the stored FireDB consensus sequences.
SQUARE assigns conservation-based reliability scores by extracting values for each aligned residue from the PSI-BLAST profiles generated from the FireDB consensus sequences. The alignment scores are smoothed with a triangular five-residue window. From the SQUARE reliability scores it is possible to discern which residues are aligned reliably, and which of the binding and functional residues are likely to have some level of functional conservation. While SQUARE was developed to evaluate the reliability of pairwise alignments, it has been shown that the method is even more effective at predicting the conservation of residues in binding sites (13). A stand-alone version of SQUARE for pairwise alignment reliability is available at http://square.bioinfo.cnio.es.
firestar
Accepted input forms are sequences in fasta format, or structures described by their PDB codes or coordinates in PDB format. Target sequences are subjected to standard PSI-BLAST searches. Profiles are generated with an nrdb90 database from the EBI (16) and the final search is made against the FireDB consensus sequence database. Functional residues mapped onto the resulting alignments and the reliability of each position in the alignment is evaluated with SQUARE. The residue scores from SQUARE represent the probability that a given target residue is aligned to the evolutionary equivalent residue in the consensus sequence. It has been shown that evolutionary conserved binding site residues are almost always involved in ligand binding in the target protein (13).
If the user input is a structure (with PDB code or uploaded structure) firestar can generate structural alignments between the query and the templates selected by PSI-BLAST with the structural alignment program LGA (17). In this case SQUARE evaluates the reliability of each position in the structural alignment.
There are three output types generated by the server:
In pairwise mode, every PSI-BLAST hit is shown and each alignment comes with the SQUARE residue-based alignment evaluation and mapping of the functional residues (Figure 1b).
In the multiple sequence mode, user selected PSI-BLAST hits are aligned with MUSCLE (18). Here the user can highlight functional residues dynamically so that comparisons between homologues are easier (Figure 1c).
In the structural alignment mode the output is based on the alignment from LGA. The output is similar to the pairwise mode, but also allows molecular visualization with a Jmol applet. The interface makes it possible to select aligned functional residues and display them in the Jmol window. This is especially valuable to observe the displacement of functional residues in homologous structures (Figure 2).
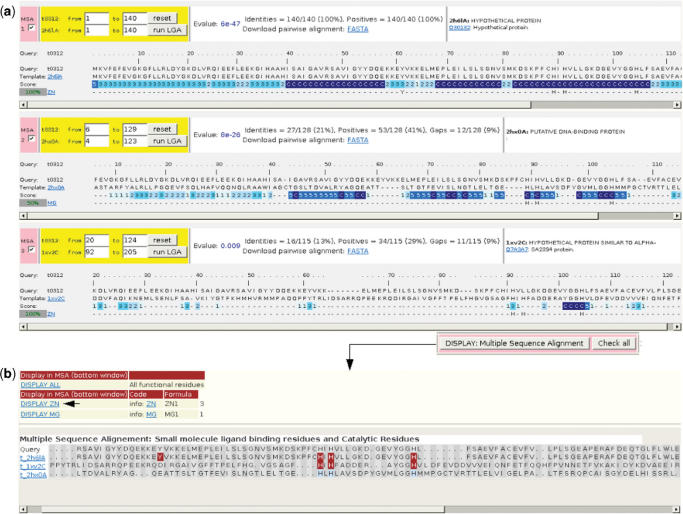
Figure 1.
firestar sequence alignment outputs. (a) The PSI-BLAST alignment between the sequence of target T0312 and the best three structures. Information about binding sites is displayed alongside the alignment reliability scores. Per residue reliability scores returned by SQUARE are in blue and are based on residue similarity and conservation of neighbouring residues as described in Tress et al. (15). (b) MUSCLE multiple sequence alignment between the query and the sequence of all three PDB templates. Functionally important residues can be displayed dynamically.
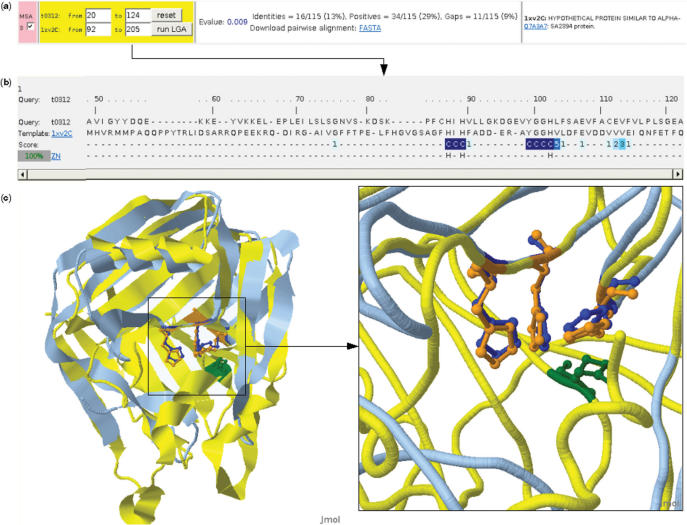
Figure 2.
firestar structural alignments and visualization. (a) Users can select the region of the structure that they wish to align. (b) The structural alignment also shows information from functional residues and the reliability of the alignment according to SQUARE. (c) The Jmol visualization window. Buttons (not shown) permit the visualization of the superpositions of each separate functional site in the two structures.
firecat—user-defined pairwise alignment inputs
The evaluation of active site conservation by SQUARE is sensitive to alignment quality, a poor alignment may mean SQUARE does not tag a binding residue as conserved. On occasions, automatic alignments may not be the most appropriate, so user-defined pairwise alignment inputs are also possible. Users may upload their own pairwise alignment with the constraint that the template they use must be in the FireDB database. FireDB is updated regularly from the PDB database, so the FireDB template database contains all the structures present in the PDB at the time of the most recent FireDB update. In this case firestar will produce an output in pairwise mode.
FUNCTIONALITY
The server interface is implemented in PHP running on apache servers. All the sequence handling, parsing and communicating with the MYSQL FireDB database is done by domestic scripts implemented in perl. firestar is integrated as part of the larger FireDB system and all outputs are cross-linked with the residue, ligand and protein information stored in FireDB. firestar is thus continually updated with the growth of the PDB and the Catalytic Site Atlas.
TESTING THE SERVER IN A PREDICTION CONTEXT. THE CASP7 FUNCTION PREDICTION EXPERIMENT
One complication with the functional data in the PDB is that bound ligands may or may not be biologically relevant. This problem was addressed in the FireDB database; ligands classified as solvent by mmCif (19) are ignored by FireDB and biological relevance can be assessed by the co-occurrence of sites in homologous proteins. Conserved sites in two or more homologues imply an evolutionary pressure in residue conservation and suggest biological relevance. firestar adds additional capacity since it makes it easier to find several homologues with same binding site conserved in aligned positions.
We developed the firestar server as one of the tools for our evaluation of the function prediction section of the 7th edition of Critical Assessment in Structure Prediction (CASP) (20). firestar was essential to determine context and conservation in other structures and whether the ligands bound to the target structures could be considered as biologically relevant or not. Only the biologically relevant ligand-binding sites formed part of the CASP evaluation.
A PRACTICAL CASE
CASP Target T0312 is a hypothetical DNA-binding protein from Archaeoglobus fulgidus. It has a homo-trimeric form in the coordinates file (PDB: 2H6L) and the three monomers bind both zinc and acetate in the same cleft. Acetate is a common solvent and it is not supposed to be relevant.
We ran firestar with the T0312 sequence, the pairwise output returned by PSI-BLAST showed two hits (Figure 1a). The first alignment in Figure 1a shows 2H6L itself and shows the zinc binding at Tyr 61 and at three conserved histidines, the second alignment is with template 2hx0 that was deposited in the PDB after the CASP7 prediction deadline. The third alignment shows the best template available to predictors, 1xv2 (21), a distant template that was found by PSI-BLAST with a high e-value and a poor alignment.
As it turns out 1xv2 also binds zinc, but only one of the three binding histidines (His 104) is reliably conserved in the PSI-BLAST alignment with the target sequence. A second histidine (His 91) is conserved but less reliable, while the third histidine and the tyrosine are not conserved. In fact two of the histidines are misaligned in the PSI-BLAST alignment, something that becomes clear in the MUSCLE alignment (Figure 1b). The histidines involved in binding in 1xv2 are also aligned in LGA structural alignment (Figure 2b) and here it can be seen that all the three histidines are also reliably aligned. Moreover the Jmol window shows that the side-chain orientations of the histidines are conserved (Figure 2c). The target clearly contains a biologically relevant zinc-binding site even though the other residues involved in zinc binding (Tyr 61 from T0312 and Glu 45 from 1xv2) were not conserved.
In addition it is clear that even though this target would be regarded as ‘difficult’, the binding residues could have been predicted by firestar before the structure was solved if the server was provided with a good alignment. The LGA structural alignment was available only because the structure has recently been released, but it would have been possible to use the firecat tool to build the correct alignment and predict the binding residues from firestar even without access to the structure.
FUTURE DIRECTIONS
Future releases of firestar will include improvements to the server interface and the development of new features. We plan to make a version available at the INB web services (http://www.inab.org/en/resources.htm), and the central web services in Canada as part of the services offered by the NoE Embrace.
We plan to exploit the predictive abilities of firestar and to make results available in the context of large annotation efforts, in particular in the BIOSAPIENS and GENEFUN projects. At the same time we are working on a version of firestar that will validate the biological relevance of all bound ligands found in PDB entries.
In addition to updating the server we are planning to test firestar with functionally interesting residues such as post-translational modifications, mutations or even residues linked to diseases in OMIM (22).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We would like to thank the valuable input and suggestions from Ana M. Rojas. This work was supported by grants: BioSapiens (LSHG-CT-2003-503265), GeneFun (LSHG-CT-2004-503567) MEC (BIO2004-00875), EMBRACE (LHSG-CT-2004-512092) and the Spanish National Bioinformatics Institute (INB, http://inab.org). Funding to pay the Open Access publication charges for this article was provided by GeneFun (LSHG-CT-2004-503567).
Conflict of interest statment. None declared.
REFERENCES
- 1.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devos D, Valencia A. Practical limits of function prediction. Proteins. 2000;41:98–107. [PubMed] [Google Scholar]
- 4.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torrance JW, Bartlett GJ, Porter CT, Thornton JM. Using a library of structural templates to recognise catalytic sites and explore their evolution in homologous families. J. Mol. Biol. 2005;347:565–581. doi: 10.1016/j.jmb.2005.01.044. [DOI] [PubMed] [Google Scholar]
- 6.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 8.Hawkins T, Luban S, Kihara D. Enhanced automated function prediction using distantly related sequences and contextual association by PFP. Prot. Sci. 2006;15:1550–1556. doi: 10.1110/ps.062153506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin DM, Berriman M, Barton GJ. GOtcha: a new method for prediction of protein function assessed by the annotation of seven genomes. BMC Bioinformatics. 2004;18:178. doi: 10.1186/1471-2105-5-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez G, Valencia A, Tress ML. FireDB–a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 2006;35:D219–D223. doi: 10.1093/nar/gkl897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tress ML, Graña O, Valencia A. SQUARE-determining reliable regions in sequence alignments. Bioinformatics. 2004;20:974–975. doi: 10.1093/bioinformatics/bth032. [DOI] [PubMed] [Google Scholar]
- 13.Tress ML, Jones DT, Valencia A. Predicting reliable regions in protein alignments from sequence profiles. J. Mol. Biol. 2003;330:705–718. doi: 10.1016/s0022-2836(03)00622-3. [DOI] [PubMed] [Google Scholar]
- 14.Westbrook J, Feng Z, Chen L, Yang H, Berman HM. The Protein Data Bank and structural genomics. Nucleic Acids Res. 2003;31:489–491. doi: 10.1093/nar/gkg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 16.Holm L, Sander C. Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics. 1998;14:423–429. doi: 10.1093/bioinformatics/14.5.423. [DOI] [PubMed] [Google Scholar]
- 17.Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acid Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourne PE, Berman HM, McMahon B, Watenpaugh KD, Westbrook J, Fitzgerald PMD. The Macromolecular Crystallographic Information File (mmCIF) Methods Enzymol. 1997;277:571–590. doi: 10.1016/s0076-6879(97)77032-0. [DOI] [PubMed] [Google Scholar]
- 20.Lopez G, Tress ML, Rojas AM, Valencia A. Assessment of predictions submitted for the CASP7 function prediction category. Proteins CASP7 Special Issue. 2007 doi: 10.1002/prot.21651. (in press) [DOI] [PubMed] [Google Scholar]
- 21.Clarke ND, Ezkurdia I, Kopp J, Read R, Schwede T, Tress ML. Domain definition and Target classification for CASP7. Proteins CASP7 Special Issue. 2007 doi: 10.1002/prot.21686. (in press) [DOI] [PubMed] [Google Scholar]
- 22.McKusick VA. Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders. 12th. Baltimore: Johns Hopkins University Press; 1998. edn. [Google Scholar]