Abstract
The identification of over-represented transcription factor binding sites from sets of co-expressed genes provides insights into the mechanisms of regulation for diverse biological contexts. oPOSSUM, an internet-based system for such studies of regulation, has been improved and expanded in this new release. New features include a worm-specific version for investigating binding sites conserved between Caenorhabditis elegans and C. briggsae, as well as a yeast-specific version for the analysis of co-expressed sets of Saccharomyces cerevisiae genes. The human and mouse applications feature improvements in ortholog mapping, sequence alignments and the delineation of multiple alternative promoters. oPOSSUM2, introduced for the analysis of over-represented combinations of motifs in human and mouse genes, has been integrated with the original oPOSSUM system. Analysis using user-defined background gene sets is now supported. The transcription factor binding site models have been updated to include new profiles from the JASPAR database. oPOSSUM is available at http://www.cisreg.ca/oPOSSUM/
INTRODUCTION
Functional genomics research often generates lists of genes with observed common properties, such as coordinated expression. For many studies, a key challenge is the generation of relevant and testable hypotheses about the regulatory networks and pathways that underlie observed co-expression. Our strategy for elucidating regulatory mechanisms identifies over-represented sequence motifs that are present in the upstream regulatory regions of genes. The motifs may represent transcription factor binding sites (TFBSs) that have a role in regulating expression.
oPOSSUM (1) and oPOSSUM2 (2) were developed to identify over-represented, predicted TFBSs and combinations of predicted TFBSs, respectively, in sets of human and mouse genes. The user inputs a list of related genes, selects the TFBS profile set to be included in the analysis, and the algorithm determines which, if any, predicted TFBSs occur in the promoters of the set of input genes more often than would be expected by chance. Both analytic approaches rely on a database of aligned, orthologous human and mouse sequences, and the delineation of conserved regions within which TFBS predictions are analyzed. While the approach does not explicitly address uncharacterized transcription factors (TFs), the effective coverage is broadened by the fact that members within certain structural families of TFs can exhibit similarities in binding specificity. While intra-class similarity is not always the case, as exemplified by the zinc-finger family of TFs (3), the observation holds true for many TF families (4,5).
Here we describe the new release of the oPOSSUM system, which integrates the two previously developed applications, and has been expanded to accommodate new species (yeast and worms). It also includes new methods for orthology assignment, transcription start site (TSS) determination and sequence alignment.
MATERIALS AND METHODS
Over-representation analysis
oPOSSUM single site analysis (SSA)
The oPOSSUM system for identifying over-represented TFBSs in sets of co-expressed genes first focused on SSA (1). Two scores were developed to assess over-representation, one at the TFBS occurrence level and the other at the gene level. The Z-score, based on the normal approximation to the binomial distribution, indicates how far and in what direction the number of TFBS occurrences deviates from the background distribution's mean. The second score, the Fisher exact test, indicates if the proportion of genes containing the TFBS is greater than would be expected by chance. TFBS predictions situated within overlapping alternative promoters are counted only once when calculating over-representation in human and mouse genes. For Caenorhabditis elegans genes in operons, TFBS predictions in the upstream region of the first gene in the operon apply to all genes in the operon.
oPOSSUM combination site analysis (CSA)
TFBSs do not act in isolation to initiate the transcription process. Transcriptional regulation can be viewed as mediated by arrays of cis-regulatory sequences, termed cis-regulatory modules (CRMs), which are bound by multiple TFs. In oPOSSUM2, Huang et al. (2) address the detection of over-represented sets of TFBSs in the promoters of a set of co-expressed genes. In brief, the method reduces combinatorial complexity through an initial clustering step, which partitions similar TFBS profiles into groups, herein denoted ‘TFBS classes’, along with an analysis step to determine a TFBS class representative profile for each TFBS class, which is used to detect over-represented sets of TFBS classes. Since each distinct, over-represented set of detected TFBS classes, herein described as a ‘TFBS class combination’, implicates the over-representation of one or more underlying TFBS profile-specific combinations, each of these TFBS class combinations is expanded to all possible TFBS profile-specific combinations (for the indicated classes) and then all combinations are analyzed for over-representation. Furthermore, given that CRMs can contain locally dense clusters of TFBSs, the system also provides for the specification of an inter-binding site distance (IBSD) constraint to confine the number of TFBS combinations that are investigated. A scoring scheme, adopted from the Fisher exact test, utilizes two sets of TFBS (class or profile-specific) combination counts to compare the degree of their over-representation: (i) the number found in the promoters of the co-expressed gene set versus (ii) the number found in the promoters of genes in a background set (all genes in the database). TFBS combinations occurring in multiple alternative gene promoter regions are counted only once.
Species-specific databases
In addition to enhancements to the human/mouse oPOSSUM database, we introduce new species databases for studies of over-represented TFBSs in yeast and worms. While the SSA over-representation analysis remains the same for all species, differences in gene structure require that the construction of the underlying databases be particular to each species.
Human/mouse
Ambiguities in ortholog assignments and the definition of TSS positions are major challenges when performing alignments for a large proportion of human and mouse genes. We have expanded the human/mouse database through (i) the discrimination of potential orthologs from predicted paralogs based on upstream sequence similarity (Figure 1), and (ii) the delineation of alternative promoters for human and mouse genes (Figure 2) to address the alignment failure observed in previous database builds.
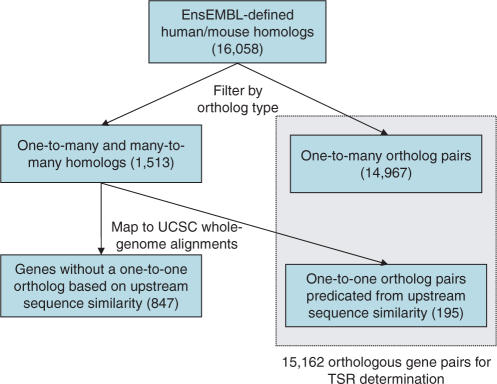
Figure 1.
Determination of one-to-one orthologs for human and mouse genes. An initial set of homologs was downloaded from EnsEMBL v41 (30). All homologs annotated as ‘one2one’ are extracted. To select the closest putative ortholog pairs from homologs with ‘one2many’ or ‘many2many’ relationships, we check for upstream conservation using the whole-genome human–mouse alignments (6). We re-annotate unambiguously aligned homologs as putative one-to-one orthologs, adding 195 gene pairs to our set and bringing the total number of orthologs to 15 162.
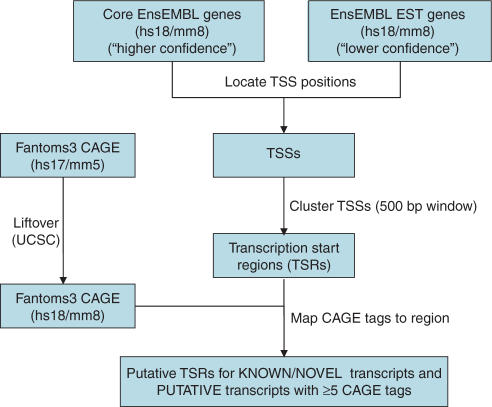
Figure 2.
Identification of transcription start regions (TSRs) using a combination of EnsEMBL annotations and CAGE data. To improve our alignments, we determine putative alternative TSSs for the human and mouse genes. For each gene, the entire repertoire of transcripts from both EnsEMBL core genes and EST genes are retrieved. The TSSs for all transcripts are recorded, followed by a clustering step such that TSSs within 500 bp of one another are merged to form a transcriptional start region (TSR). For each TSR containing a transcript annotated as ‘known’ or ‘novel’, we accept the TSR as is. For TSRs based solely on EST gene transcripts, we require a minimum of 5 CAGE tags as evidence for transcription initiation.
While the inclusion of promoter comparisons for candidate ortholog assignment may be controversial, the impact is marginal as <1.3% of gene pairs were derived from this approach. This brings the total number of orthologs to 15 162. Despite improvements in EnsEMBL's ortholog prediction, this is only 1079 more orthologs than were present in our previous database build. Based on the small incremental increases in mapped orthologs, we may be nearing the upper bound for the number of genes in human and mouse that are truly orthologous and detectable by sequence conservation. Detailed descriptions of transcription start region (TSR) determination and the distribution of TSRs for human and mouse genes are available as Supplementary Data.
For each human/mouse orthologous pair, we determine the coordinates of the longest region from the UCSC genome alignments (6) spanning all transcripts plus an additional 10 kb of upstream sequence. The orthologous sequences are retrieved and re-aligned using ORCA, a pairwise global progressive alignment algorithm (1) to optimally align short, conserved blocks within longer global alignments. If possible, TSRs from human and mouse are paired in the alignment. We apply three dynamically computed and progressively more stringent conservation thresholds corresponding to the top 10, 20 and 30% of all 100-bp non-coding windows, each with a minimum percent identity of 70, 65 and 60%, respectively. Of the 15 162 orthologous gene pairs supplied as input to the oPOSSUM pipeline, 15 121 (99.7%) successfully align, and 15 027 (99.1%) have non-exonic conserved regions above 60% nucleotide identity. This is a significant improvement over the previous version of oPOSSUM.
Caenorhabditis elegans/Caenorhabditis briggsae
To facilitate transcriptional regulatory analysis of the numerous gene expression studies performed in C. elegans, we have implemented a worm version of oPOSSUM. While the database structure and pipeline procedure are very similar to that used for the human/mouse database, there are small modifications that allow for mapping of genes to their operons, as defined by Blumenthal et al. (7). In addition, nucleotide identity thresholds for conserved regions were reduced to 60, 55 and 50% for the top 10, 20 and 30% of non-coding windows, respectively, to account for the greater sequence divergence between C. elegans and C. briggsae compared to human and mouse. The set of orthologs for C. elegans and C. briggsae is defined by one-to-one InParanoid clusters (8) from WormBase (WS160) (9). After filtering overlapping genes, 10592 orthologous gene pairs (of which, 2140 genes are in operons) remain for alignment. Alignments are performed on the orthologous gene sequences plus 2 kb of upstream sequence (relative to the start codon) for C. elegans, and 4 kb of upstream sequence for C. briggsae. Annotations are not as mature for C. briggsae, and the longer upstream region aids in the alignment of the worm promoter sequences. Alternative promoters have not been considered in this first version; however, should CAGE data or other reliable means for annotating TSSs in worms become available, efforts will certainly be made to include them. Of the 10 592 worm orthologs, 9331 (88%) successfully align.
Yeast
The analysis of yeast promoters is simplified by the more compact nature of the yeast genome. This characteristic diminishes the requirement for comparative methods to reduce the search space and noise inherent in larger genomes. Computational methods using S. cerevisiae sequences alone have successfully been used to identify regulatory elements associated with known sets of related genes (10,11). We opted to exclude phylogenetic footprinting for yeast, and instead, select promoter sequences corresponding to the 5′ untranslated region 1000 bp immediately upstream of the start codon of each open reading frame (ORF). Note that for all applications, users have the option to further restrict the search space if they wish. The sequences were downloaded from the Saccharomyces Genome Database (12).
TFBS prediction
For the metazoan species, we search for matches to TFBS profiles contained in the JASPAR CORE and JASPAR PhyloFACTS database collections (13,14). Additionally, we include a set of profiles compiled for C. elegans TFs from literature review for Worm SSA (Table S2). Binding sites are predicted for the sequences using the TFBS suite of Perl modules for regulatory sequence analysis (15). A predicted binding site for a given TF model is reported if the site occurs in the promoters of both orthologs above a threshold PSSM score of 70% and at equivalent positions in the alignment. Overlapping sites for the same TF are filtered such that only the highest scoring motif is kept. The genomic location, profile score, motif orientation and local sequence conservation level of each TFBS match in orthologous genes are stored in the respective species databases. For S. cerevisiae, we compiled a collection of yeast-specific TFBS motifs from both the Yeast Regulatory Sequence Analysis (YRSA) system (16) and the literature (Table S3), and record the genomic location, profile score and motif orientation for each prediction.
Based on the observation that members of the same structural family of TFs often bind to similar sequences, plant and insect matrices are available for inclusion in the analysis. The MADS family of TFs is an excellent example of conservation of binding domains between plants and vertebrates (17,18), and there are numerous examples of conservation of binding domains across vertebrates, flies and worms. Thus, in cases where a profile for the TF of interest is not available in the database, oPOSSUM can still provide insights into the underlying regulation by suggesting a particular TF family that may be involved.
RESULTS AND DISCUSSION
Examples of applications
Each oPOSSUM component was validated on sets of reference genes. The results of all validations are available as Supplementary Data (Tables S4–S13). In the interest of space, selected examples are described for each system.
Human SSA
Wonsey and Follettie (19) performed a microarray analysis of genes that are transcriptionally regulated by FoxM1, a member of the forkhead family of TFs, using BT-20 cells that had been transfected with FoxM1 siRNA. They identified a set of 27 genes that were specifically regulated in cells transfected with FoxM1 siRNA (Table S4). The 27 Affymetrix UG144A identifiers were mapped to 27 EnsEMBL gene identifiers and submitted to Human SSA with default parameters. Of these, 22 genes had a unique mouse ortholog and were used in the oPOSSUM analysis. While a specific profile for FoxM1 is not present in JASPAR CORE, other members of the forkhead family were ranked in the top 10 highest scoring TFBS profiles (Table 1). There is also a known association between HNF4, the highest scoring TFBS profile, and the forkhead TF, FOXO1 in the regulation of gluconeogenic gene expression in hepatocytes (20), which may explain the over-representation of the HNF4 profile.
Table 1.
oPOSSUM results for human FoxM1-regulated gene cluster
| JASPAR CORE | TF Class | IC | Target gene hits | Background TFBS rate | Target TFBS rate | Z-score | Fisher score |
|---|---|---|---|---|---|---|---|
| HNF4 | Nuclear | 9.62 | 13 | 0.0054 | 0.0085 | 7.19 | 2.64E−02 |
| Fos | bZIP | 10.67 | 15 | 0.0111 | 0.0146 | 5.72 | 4.29E−01 |
| Pbx | Homeo | 14.64 | 5 | 0.0019 | 0.0033 | 5.57 | 3.10E−01 |
| FOXI1 | Forkhead | 13.18 | 16 | 0.0153 | 0.0186 | 4.49 | 9.05E−02 |
| RORA1 | Nuclear Receptor | 17.42 | 4 | 0.0020 | 0.0029 | 3.54 | 5.04E−01 |
| TAL1-TCF3 | bHLH | 14.07 | 12 | 0.0052 | 0.0066 | 3.30 | 5.88E−02 |
| Staf | Zn-Finger, C2H2 | 17.54 | 3 | 0.0014 | 0.0021 | 3.16 | 3.03E−01 |
| Foxa2 | Forkhead | 12.43 | 13 | 0.0152 | 0.0174 | 3.04 | 4.83E−01 |
| Foxd3 | Forkhead | 12.94 | 13 | 0.0172 | 0.0194 | 2.93 | 5.27E−01 |
| TEAD | TEA | 15.67 | 6 | 0.0028 | 0.0037 | 2.85 | 4.70E−01 |
We previously identified over-represented Fos binding sites in a set of genes induced after transformation by c-Fos in rat fibroblast cells (1,21). We analyzed 160 orthologous genes from the original list of 252 induced genes (Table S7). This is a notable improvement over the previous version where only 98 genes were included in the oPOSSUM analysis. The Fos TFBS profile ranked second in the list of over-represented TFBSs (Table s7). Inspection of the results using the JASPAR PhyloFACTS profiles with default parameters illustrates how inclusion of this new set of profiles provides additional, meaningful information (Table 2). The highest ranked PhyloFACTS motif (TGANTCA) is noted by JASPAR as being most similar to the binding profile for AP-1, and the third highest scoring motif (TGASTMAGC) is most similar to the bZIP TF NF-E2. AP-1 complexes are comprised of Fos and Jun proteins, and the structurally related NF-E2 and AP-1 TFs bind similar sequence motifs (22).
Table 2.
oPOSSUM results for c-Fos-regulated gene cluster
| JASPAR PhyloFACTS | Similar to | IC | Target gene hits | Background TFBS rate | Target TFBS rate | Z-score | Fisher score |
|---|---|---|---|---|---|---|---|
| TGANTCA | AP-1 | 12.06 | 46 | 0.0011 | 0.0023 | 18.05 | 1.40E−04 |
| GGGYGTGNY | – | 14.18 | 82 | 0.0059 | 0.0083 | 15.64 | 4.98E−02 |
| TGASTMAGC | NF-E2 | 16.60 | 43 | 0.0013 | 0.0024 | 15.64 | 1.19E−03 |
| GGARNTKYCCA | – | 17.13 | 44 | 0.0016 | 0.0026 | 12.54 | 1.11E−03 |
| GGGAGGRR | MAZ | 14.00 | 111 | 0.0171 | 0.0202 | 11.98 | 3.16E−01 |
Human CSA
The CSA was validated on a set of mouse skeletal muscle genes comprised of the union of the results of the microarray studies of Moran et al. (23) and Tomczak et al. (24) (Table S9). To avoid circularity, we removed muscle-specific genes used to generate the JASPAR binding site profiles for Mef-2, Myf, Sp-1, SRF and Tef. These factors occur in clusters in CRMs that contribute to skeletal muscle-specific expression (25). Table 3 lists the top five over-represented pairwise TFBS combinations for this set of genes, along with the JASPAR class each TF profile clustered to, and the Fisher score obtained for each pair. The five most over-represented pairs of TFBS profiles include combinations of Mef-2, SRF and Sp-1.
Table 3.
oPOSSUM results for skeletal muscle genes identified by Moran et al. and Tomczak et al
| TF name (Class ID) | TF class name | TF name (Class ID) | TF class name | Score |
|---|---|---|---|---|
| MEF2A (class 4) | MADS | Myf (class 22) | bHLH | 1.65E−06 |
| MEF2A (class 4) | MADS | ZNF42_1–4 (class 25) | Zn-finger, C2H2 | 4.24E−06 |
| Myf (class 22) | bHLH | SRF (class 1) | MADS | 2.52E−05 |
| SP1 (class (31) | Zn-finger, C2H2 | SRF (class 1) | MADS | 2.68E−05 |
| Agamous (class 1) | MADS | MEF2A (class 4) | MADS | 7.63E−05 |
The inclusion of alternative promoters provides notable improvements in the Human SSA and Human CSA analyses. The same datasets were used to validate our previous and current human oPOSSUM analyses systems. Demarcation of additional promoter boundaries increases the signal in the discovery process, improving the signal for both over-represented single TFBSs and combinations of TFBSs in the gene sets analyzed.
Worm SSA
Worm SSA was tested on a set of well-characterized nematode muscle genes (Table S10) (26). Analysis of 1000 bp of upstream sequence, using the top 10% of conserved regions (minimum of 60% sequence identity), a matrix match threshold of 80% and the worm profiles, identified the putative muscle1 motif with a Z-score of 20.6 and a Fisher score <0.01 (Table 4). This is, however, somewhat circular, given that 19 of the 41 input genes were used to generate the putative muscle-specific worm profiles. Analysis using the JASPAR CORE profiles ranked SP1 and Su(H) within the top 10 scoring profiles (Table S10B). Studies in Xenopus and Drosophila provide evidence that MyoD triggers Notch signaling through Su(H) for muscle determination (27,28). Although SP1 has been implicated in muscle CRMs, it is a general TF involved in the expression of many different genes and binds to GC-rich motifs.
Table 4.
oPOSSUM results for worm skeletal muscle genes using worm profiles
| Worm | Status | IC | Target gene hits | Background TFBS rate | Target TFBS rate | Z-score | Fisher score |
|---|---|---|---|---|---|---|---|
| Muscle1 | Putative | 11.34 | 6 | 0.0025 | 0.0156 | 20.56 | 4.24E–04 |
| Muscle2 | Putative | 11.97 | 4 | 0.0022 | 0.0089 | 11.19 | 1.39E−02 |
| LIN-14 | Putative | 9.13 | 9 | 0.0143 | 0.0280 | 9.12 | 1.17E−01 |
| Muscle3 | Putative | 16.67 | 4 | 0.0029 | 0.0064 | 5.02 | 6.96E−02 |
Yeast SSA
The yeast CLB2 gene cluster is comprised of 32 genes whose pattern of expression peaks at late G2/early M phase of the cell cycle (Table S11). Transcription of these genes is regulated by two TFs: FKH, which is a component of the TF SFF, and MCM1, a member of the early cell cycle box (ECB) binding complex. Analysis of 500 bp of upstream sequence using a matrix match threshold of 85% ranked ECB, MCM1 and FKH1 in the top five scoring TFBS profiles (Table 5), which is consistent with the literature (29).
Table 5.
oPOSSUM results for the yeast CLB2 gene cluster
| YEAST | TF Class | IC | Target gene hits | Background TFBS rate | Target TFBS rate | Z-score | Fisher score |
|---|---|---|---|---|---|---|---|
| ECB | Unclassified | 16.65 | 13 | 0.0019 | 0.0131 | 32.87 | 8.68E−09 |
| MCM1 | MADS | 9.15 | 10 | 0.0073 | 0.0165 | 13.71 | 1.08E−02 |
| FKH1 | Forkhead | 13.28 | 30 | 0.0305 | 0.0473 | 12.26 | 4.05E−02 |
| CCA | Unclassified | 16.93 | 3 | 0.0017 | 0.0040 | 7.08 | 2.02E−01 |
| LYS14 | C6_Zinc finger | 17.02 | 6 | 0.0030 | 0.0053 | 5.20 | 9.41E−02 |
Web server
The four oPOSSUM systems, Human SSA, Human CSA, Worm SSA and Yeast SSA, have been integrated into a use-friendly website at: http://www.cisreg.ca/oPOSSUM/. We recommend that users of the system begin with the SSA to quickly identify TFBSs that may be relevant to their input data sets. For sets of human and mouse genes, this can be followed with the CSA, which takes longer to process, but which can provide insights into TFBSs that may be acting in concert to regulate the set of genes.
The web implementation allows for analysis in default and custom modes. Default mode processing is faster as TFBS counts have been pre-calculated and stored for pre-defined conservation levels, matrix match thresholds and promoter lengths. In either mode, the user is required to select a species and to enter a list of gene identifiers (EnsEMBL, RefSeq, HGNC and Entrez Gene are supported for human). A number of options are available to specify the TFBS profile set to be used in the analysis. Finally, the conservation level, matrix match threshold and the promoter length can be varied. In the custom mode, users may define their own background set, which provides users with more control, but results in more variable processing speeds depending on the size of the background set and the parameters selected.
Upon submission, oPOSSUM SSA generates a summary of the input parameters, and produces a single table that ranks the over-represented TFBSs by descending Z-score. The table may be sorted by TF name, TF class, supergroup, information content (IC), Z-score and Fisher score (Figure 3A). Pop-up windows linked to each TFBS foreground count display the genes in which the putative site is located, the promoter region(s) for each gene, as well as the TFBS's co-ordinates and score (Figure 3B). TFBSs that occur in overlapping promoter regions are marked by an asterisk and highlighted in yellow. The TF names are linked to the JASPAR database for easy access to information regarding the binding site profiles. The output for oPOSSUM CSA is similar, providing (i) a ranked list of over-represented TFBS class combinations, and (ii) a list of the most significant TFBS combinations (found in the set of expanded top-ranked class combinations).
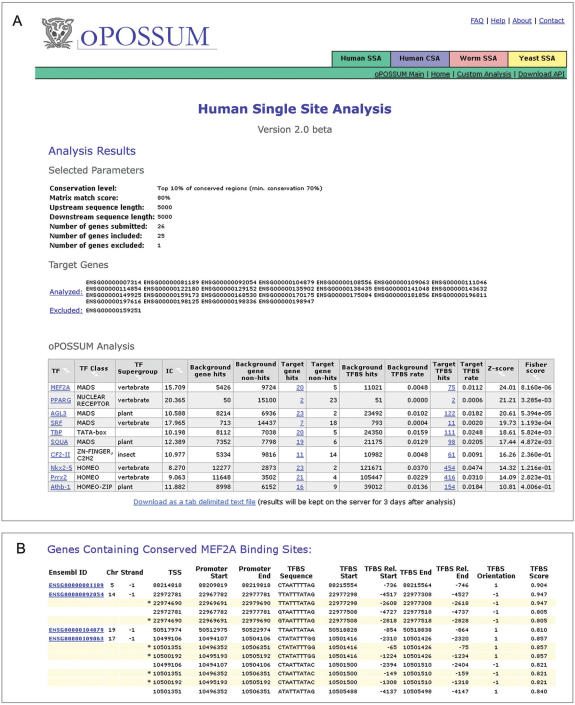
Figure 3.
(A) A screenshot of the output of the oPOSSUM Human SSA analysis, with TFBS profiles ranked by Z-score. The arrows allow the user to sort and re-order the results by Fisher score, TF name, TF class, TF supergroup or TF profile information content (IC). Each TF name links to a pop-up window displaying the TFBS profile information. (B) Pop-up window displaying genes that contain a particular TFBS (in this case, MEF2A; partial list shown), as well as the promoter coordinates associated with each gene, and the motif locations and scores. Sites in overlapping alternative promoters are highlighted for emphasis. Such sites are only counted once in the statistical analysis.
Based on the underlying assumption of the statistics employed that DNA sequences are randomly generated, there is little reason to accept the calculated scores as accurate reflections of significance. Instead, as suggested in the original published description of the oPOSSUM algorithm, we recommend that the scores are best used as rankings rather than significance measures. For this reason, a multiple testing correction is not applied as it does not alter the relative ranks. Empirically, we determined that TFBS profiles with Z-scores ⩾10 and Fisher scores ⩽0.01 facilitate the identification of relevant TFBSs for our sets of reference genes (1). However, these are relatively stringent thresholds, and we encourage users to examine the scores of top-ranked TFBS profiles before applying any cutoffs.
We provide a consistent display for all four systems. However, there are slight differences between the systems, such as different parameters for selection on the input pages which are relevant for each species database and system. Also, due to the longer processing times required to compute combinations of TFBSs, Human CSA queues the analysis request on the server and emails the completed results to the user.
CONCLUSIONS
The oPOSSUM system is under continued development. Efforts are underway to allow users to submit custom TF profiles to be included in the analysis. An improved search method for nuclear hormone receptors, which typically contain two half sites separated by a variable length spacer, has been developed and will be included in a future release. We will continue to add TFBS profiles as they become available, with an emphasis on expanding the repertoire of worm TFBS profiles. We believe the oPOSSUM web server is and will continue to be a useful resource for inference of mechanisms of co-regulation based on observed co-expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Project funding provided through the Canadian Institutes of Health Research (CIHR) and Merck. A.T.K. and S.J.H.S. are supported by the National Science and Engineering Research Council of Canada (NSERC); D.L.F., S.J.H.S. and W.W.W. are supported by the Michael Smith Foundation for Health Research (MSFHR); and D.L.F. and W.W.W. are supported by the CIHR. The authors would like to thank Jonathan Lim and Miroslav Hatas for systems support. The Open Access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang SS, Fulton DL, Arenillas DJ, Perco P, Ho Sui SJ, Mortimer JR, Wasserman WW. Advances in Bioinformatics and Computational Biology. Vol. 3. London, UK: Imperial College Press; 2006. Identification of over-represented combinations of transcription factor binding sites in sets of co-expressed genes; pp. 247–256. [Google Scholar]
- 3.Urnov FD, Rebar EJ. Designed transcription factors as tools for therapeutics and functional genomics. Biochem. Pharmacol. 2002;64:919–923. doi: 10.1016/s0006-2952(02)01150-4. [DOI] [PubMed] [Google Scholar]
- 4.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1:REVIEWS001. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, et al. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang MQ. Promoter analysis of co-regulated genes in the yeast genome. Comput. Chem. 1999;23:233–250. doi: 10.1016/s0097-8485(99)00020-0. [DOI] [PubMed] [Google Scholar]
- 11.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat. Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 12.Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenhard B, Wasserman WW. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 16.Sandelin A, Hoglund A, Lenhard B, Wasserman WW. Integrated analysis of yeast regulatory sequences for biologically linked clusters of genes. Funct. Integr. Genomics. 2003;3:125–134. doi: 10.1007/s10142-003-0086-6. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, Ribas DP, Martinez-Castilla L, Yanofsky MF. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl Acad. Sci. USA. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 19.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J. Biol. Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 21.Ordway JM, Williams K, Curran T. Transcription repression in oncogenic transformation: common targets of epigenetic repression in cells transformed by Fos, Ras or Dnmt1. Oncogene. 2004;23:3737–3748. doi: 10.1038/sj.onc.1207483. [DOI] [PubMed] [Google Scholar]
- 22.Daftari P, Gavva NR, Shen CK. Distinction between AP1 and NF-E2 factor-binding at specific chromatin regions in mammalian cells. Oncogene. 1999;18:5482–5486. doi: 10.1038/sj.onc.1202916. [DOI] [PubMed] [Google Scholar]
- 23.Moran JL, Li Y, Hill AA, Mounts WM, Miller CP. Gene expression changes during mouse skeletal myoblast differentiation revealed by transcriptional profiling. Physiol. Genomics. 2002;10:103–111. doi: 10.1152/physiolgenomics.00011.2002. [DOI] [PubMed] [Google Scholar]
- 24.Tomczak KK, Marinescu VD, Ramoni MF, Sanoudou D, Montanaro F, Han M, Kunkel LM, Kohane IS, Beggs AH. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18:403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- 25.Wasserman WW, Fickett JW. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- 26.GuhaThakurta D, Schriefer LA, Waterston RH, Stormo GD. Novel transcription regulatory elements in Caenorhabditis elegans muscle genes. Genome Res. 2004;14:2457–2468. doi: 10.1101/gr.2961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusconi JC, Corbin V. Evidence for a novel Notch pathway required for muscle precursor selection in Drosophila. Mech. Dev. 1998;79:39–50. doi: 10.1016/s0925-4773(98)00170-1. [DOI] [PubMed] [Google Scholar]
- 28.Wittenberger T, Steinbach OC, Authaler A, Kopan R, Rupp RA. MyoD stimulates delta-1 transcription and triggers notch signaling in the Xenopus gastrula. EMBO J. 1999;18:1915–1922. doi: 10.1093/emboj/18.7.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreiman G. Identification of sparsely distributed clusters of cis-regulatory elements in sets of co-expressed genes. Nucleic Acids Res. 2004;32:2889–2900. doi: 10.1093/nar/gkh614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.