Abstract
PAR-3D (http://sunserver.cdfd.org.in:8080/protease/PAR_3D/index.html) is a web-based tool that exploits the fact that relative juxtaposition of active site residues is a conserved feature in functionally related protein families. The server uses previously calculated and stored values of geometrical parameters of a set of known proteins (training set) for prediction of active site residues in a query protein structure. PAR-3D stores motifs for different classes of proteases, the ten glycolytic pathway enzymes and metal-binding sites. The server accepts the structures in the pdb format. The first step during the prediction is the extraction of probable active site residues from the query structure. Spatial arrangement of the probable active site residues is then determined in terms of geometrical parameters. These are compared with stored geometries of the different motifs. Its speed and efficiency make it a beneficial tool for structural genomics projects, especially when the biochemical function of the protein has not been characterized.
INTRODUCTION
Increasing structural genomics projects have led to the exponential growth of the number of available protein structures. A few of these structures are annotated as hypothetical proteins as biochemical information is not available for them. Experimental functional characterization of proteins is a labor expensive and time consuming process. A computational tool is therefore useful to predict the functional site in a protein. The importance of such a tool is strengthened by the automation required for structural genomics projects.
A large number of theoretical tools exist that attempt to predict functions of proteins on the basis of sequence or structural homology of the query protein with well-characterized proteins. However, proteins having sequence or structural similarity might not always perform similar biological functions (1,2). Proteins possessing different folds are also known to perform similar functions such as subtilisin-like proteases and trypsin-like proteases (3). This discrepancy has led to the development of structure-based approaches wherein function is predicted on the basis of similarity of the spatial arrangement of functionally significant residues.
Structure-based approaches (3–9) typically attempt to identify residues that might be non-contiguous in the primary sequence but are structurally analogous with a known structural template. Such approaches are guided by the fact that proteins perform similar function by maintaining the physicochemical environment of their functionally significant residues. This fact can be exploited to generate structural templates from active site geometries of known enzymes, and then comparing the newly determined structures with these templates. Methods that create structural templates from Cα atoms of the active site residues and their spatial neighbors have a drawback of lacking specificity and thereby giving rise to a large number of false positives. The other methods that use all the side-chain atoms of the key residues are too constrained and often overlook the small variations that might occur in the side chain placements. In our method, we use Cβ atoms of the key residues along with the corresponding Cα atoms to form a template. These templates possess optimum specificity and flexibility to identify active site residues in query structures.
The current method is highly specific for each functional class of proteins included here. The method is not affected by the small conformational variations and ambiguities in the placement of side-chains in the query structure. In addition to this the algorithm employed does not require any similarity to overall sequence or fold of known proteins. In this article we describe a web-server that executes this structure-based approach for predicting function.
IMPLEMENTATION
The method of Iengar and Ramakrishnan (10) has been modified and implemented in the current server. Structural templates are generated for the active site residues of different protease classes, glycolytic pathway enzymes and metal-binding sites. A training set was formulated from a set of known proteins of each functional family. The structural templates consist of active site residues’ identity and the geometrical parameters derived from their spatial environment. The geometrical parameters considered are the distances between the Cα and Cβ atoms of the active site residues and the angle between the Cα plane and the Cβ plane. The Cα and Cβ planes are defined by the Cα or Cβ atoms, respectively of the residues comprising the active site (Figure 2a). Structural templates derived for proteases also considered the primary sequence order. Geometrical parameters for all the structural motifs are calculated and stored for the prediction (http://sunserver.cdfd.org.in:8080/protease/PAR_3D/motif.html).
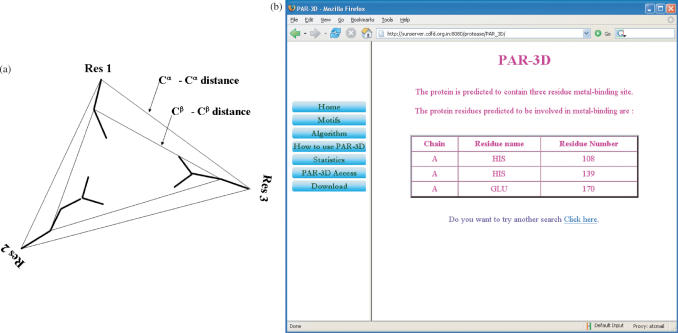
Figure 2.
(a) Typical protease class structural template. The seven parameters used to define the template are distances between three Cα atoms, distances between three Cβ atoms and the angle between the planes formed by the three Cα and the three Cβ atoms of the active site residues. (b) Output of a search carried out for yeast YDR533c structure (1QVV) shows a putative metal-binding site predicted by PAR-3D.
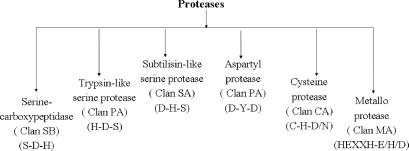
MEROPS database (11,12) was used to form training sets for different protease classes. MEROPS classifies proteases into 47 different clans on the basis of their evolutionary origin. Structural templates could be generated for six of these clans. The clan identifiers along with the active site residue pattern are shown (Figure 1). Templates could not be generated for other clans either due to non-availability of sufficient representative structures or due to involvement of less than three residues in the catalytic activity.
Figure 1.
The flow chart displays structural templates generated for different protease classes. Structural templates represent six clans described in the MEROPS database. The clan identifier and the primary sequence order of their active site residues are also shown.
The algorithm employed here performs well for all the structures solved by X-ray crystallography, NMR spectroscopy and theoretical structure prediction tools. In the case of NMR structures only first three models are considered for the prediction of functional site residues. However, the server is especially useful for structures modeled by threading, because inaccurate side chain placement in the threading-based models does not affect the accuracy of active-site residue prediction.
A two-step procedure is used to identify active site residues for every query structure. In the first step coordinates of residues that can form the active site are extracted from the query structure file. In the second step spatial arrangement of the probable active site residues is determined in terms of geometrical parameters. These parameters are compared with the stored geometries of different functional classes.
INPUT AND OUTPUT
The user is required to provide a single query structure file in a PDB format. The user submitted structure files are accepted through an HTML form generated using CGI-Perl script. In order to search for a RCSB file, it has to be downloaded on a local machine and then submitted to PAR-3D. PAR-3D stores structural motifs for different protease classes, glycolytic pathway enzymes and metal-binding sites. The user can specify if they wish to search against one family of motifs or all the PAR-3D motifs. The uploaded file is first tested and verified for the PDB format. The structure data obtained are then processed by a set of PERL scripts that search for the stored structural templates.
Output is provided in a tabular format describing the list of predicted active site residues. A sample output produced using yeast YDR533c structure (PDB ID: 1QVV) is shown in Figure 2b. The first column lists the chain identifier. The second column provides residue name and the third column lists the residue number as defined in the uploaded file. The structural motifs stored here for the comparison are specific for different protease classes, glycolytic pathway enzymes and metal-binding sites. Therefore, output also provides information about the functional class of the predicted site in the query structure.
LIMITATION
Algorithm implemented here predicts the functional class of the query structure on the basis of the spatial arrangement and the residue identity of the predicted catalytic residues. However, it is known that several functional classes belonging to the superfamily hydrolases, such as acetyl-choline esterases, acetonitriles, lipases, serine carboxypeptidases, all possess similar catalytic triads as serine carboxypeptidases due to similar catalytic mechanism. Therefore, a query structure from any of these hydrolases will be predicted as serine carboxypeptidases.
PERFORMANCE AND AVAILABILITY
The server (PAR-3D) has been tested extensively and response time has been observed to be 1–2 min. The local running time of the same was 0.3 CPU seconds on a silicon graphics workstation with R10000 processor. Server has been used to scan the complete PDB database. The statistics of PAR-3D performance upon scanning the complete PDB can be accessed at http://sunserver.cdfd.org.in:8080/protease/PAR_3D/statistics.html.
The PAR-3D web server is freely available at http://sunserver.cdfd.org.in:8080/protease/PAR_3D/index.html. It can be accessed through a browser using any operating system.
CONCLUSION AND FUTURE WORK
PAR-3D web server identifies active site residues in the query structure using structural motifs. The algorithm used for the server has been used to scan the entire PDB. Presently the server searches for structural motifs derived from proteases, glycolytic pathway enzymes and metal-binding sites. We are currently working to generate structural motifs for other functionally important sites in proteins. We are also working to include a feature, which will help to engineer new catalytic sites in existing proteins. With the availability of structural motif for several functional classes of proteins, this tool will be beneficial for structural genomics projects.
ACKNOWLEDGEMENTS
We acknowledge the overall support of S. K. Basu, S. E. Hasnain and J. Gowrishankar in this work. The support of National Institute of Immunology SUN Centre of Excellence in Medical Bioinformatics, CDFD for providing computational resources is gratefully acknowledged. K.G. is a CSIR Senior Research Fellow. S.C.M. is a Wellcome Trust International Senior Research Fellow. Funding to pay the Open Access publication charges for this article was provided by WT070006.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12:1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 3.Wallace AC, Laskowski RA, Thornton JM. Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci. 1996;5:1001–1013. doi: 10.1002/pro.5560050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artymiuk PJ, Poirrette AR, Grindley HM, Rice DW, Willett P. A graph-theoretic approach to the identification of three-dimensional patterns of amino acid side-chains in protein structures. J. Mol. Biol. 1994;243:327–344. doi: 10.1006/jmbi.1994.1657. [DOI] [PubMed] [Google Scholar]
- 5.Fetrow JS, Siew N, Skolnick J. Structure-based functional motif identifies a potential disulfide oxidoreductase active site in the serine/threonine protein phosphatase-1 subfamily. Faseb J. 1999;13:1866–1874. doi: 10.1096/fasebj.13.13.1866. [DOI] [PubMed] [Google Scholar]
- 6.Fischer D, Wolfson H, Lin SL, Nussinov R. Three-dimensional, sequence order-independent structural comparison of a serine protease against the crystallographic database reveals active site similarities: potential implications to evolution and to protein folding. Protein Sci. 1994;3:769–778. doi: 10.1002/pro.5560030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milik M, Szalma S, Olszewski KA. Common structural cliques: a tool for protein structure and function analysis. Protein Eng. 2003;16:543–552. doi: 10.1093/protein/gzg080. [DOI] [PubMed] [Google Scholar]
- 8.Tendulkar AV, Wangikar PP, Sohoni MA, Samant VV, Mone CY. Parameterization and classification of the protein universe via geometric techniques. J. Mol. Biol. 2003;334:157–172. doi: 10.1016/j.jmb.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Wangikar PP, Tendulkar AV, Ramya S, Mali DN, Sarawagi S. Functional sites in protein families uncovered via an objective and automated graph theoretic approach. J. Mol. Biol. 2003;326:955–978. doi: 10.1016/s0022-2836(02)01384-0. [DOI] [PubMed] [Google Scholar]
- 10.Iengar P, Ramakrishnan C. Knowledge-based modeling of the serine protease triad into non-proteases. Protein Eng. 1999;12:649–656. doi: 10.1093/protein/12.8.649. [DOI] [PubMed] [Google Scholar]
- 11.Rawlings ND, Tolle DP, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlings ND, Morton FR, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]