Abstract
Clustered regularly interspaced short palindromic repeats (CRISPRs) constitute a particular family of tandem repeats found in a wide range of prokaryotic genomes (half of eubacteria and almost all archaea). They consist of a succession of highly conserved regions (DR) varying in size from 23 to 47 bp, separated by similarly sized unique sequences (spacer) of usually viral origin. A CRISPR cluster is flanked on one side by an AT-rich sequence called the leader and assumed to be a transcriptional promoter. Recent studies suggest that this structure represents a putative RNA-interference-based immune system. Here we describe CRISPRFinder, a web service offering tools to (i) detect CRISPRs including the shortest ones (one or two motifs); (ii) define DRs and extract spacers; (iii) get the flanking sequences to determine the leader; (iv) blast spacers against Genbank database and (v) check if the DR is found elsewhere in prokaryotic sequenced genomes. CRISPRFinder is freely accessible at http://crispr.u-psud.fr/Server/CRISPRfinder.php.
INTRODUCTION
Genomic structures corresponding to CRISPRs were observed first in 1987 in Escherichia coli (1) and were subsequently reported in other organisms under different names [TREP (2), SRSR (3,4), DRVs (5), LCTR (6), SPIDR (7)] until the CRISPR acronym was proposed by Jansen et al. (8). The direct repeat sequences carry in general a low level of palindromic symmetry; they are remarkably well conserved within a species (up to 248 exact copies in Verminephrobacter eiseniae EF01-2). However, one of the flanking DRs is frequently truncated or diverged (see Supplementary Data). The DR size varies from 24 to 47 bp whereas the spacer sequence is generally within the range of 0.6–2.5× the DR size. The originality of spacers is that they apparently derive from conjugative plasmids or bacteriophages (2,9–11). A prokaryotic genome may harbour up to 16 CRISPR clusters with the same or a different DR. In a genome, a single CRISPR is generally associated with a family of genes called cas for CRISPR-associated (8,12), encoding proteins showing functional similarity with components of the eukaryotic RNA interference (RNAi) systems (13). In addition, it was demonstrated in two archaea, Archaeoglobus fulgidus (14) and Sulfolobus solfataricus (15), that the CRISPR locus is transcribed into small RNAs (smRNA) probably from one of the flanking regions, the leader, acting as a promoter. These observations and the viral origin of spacers have led to the hypothesis that the CRISPR-associated system (CASS) is a prokaryotic defence mechanism against genetic aggressions (10,13,16). Within species, CRISPRs may be present in a subset of strains, where they sometimes show polymorphism. The DR and the order of the spacers are well conserved, but the number of motifs (DR + spacer) differs from strain to strain. To better understand the mechanisms underlying the CRISPRs’ evolutionary scenario, three evolution rules were proposed by Pourcel et al. (10) and confirmed by Lillestol et al. (15): (i) polarized acquisition of spacers near the leader sequence; (ii) random loss of motifs and (iii) shared ancestry when spacers are identical.
CRISPRs’ in silico analyses started in 1995 (2) but no specific stand-alone CRISPR software tool was created. Several software were used by different authors to identify these particular repeats but usually a manual discard of background was necessary, and generally some CRISPR clusters were missed or neglected, especially the shortest one (less than three motifs). This is the case, for example, of Tandem Repeat Finder (17) when considering a motif (DR + spacer) as a degenerate repeat (10,18), or Locating Uniform poly-Nucleotide Areas (LUNA), a program for finding degenerate repeats in microbial genomes on a desktop computer. The repeats can be filtered using several parameters including length, distance and level of conservation. LUNA was used especially for finding CRISPRs in archaea (4,15). Another program, Patscan (19) a pattern-matching tool that searches sequences fitting the introduced pattern, was applied to identify CRISPRs containing at least three (20) or four exact direct repeats (8). PYGRAM (21) is a visualization program browsing all the repeats in the submitted genomic sequence and showing perfectly conserved palindromic repeats as pyramids. The PYGRAM program is mostly efficient in visually displaying large CRISPRs (CRISPRs with as many as seven motifs are considered as being very short in this work) since they will be recognized as a concentration of horizontal bars referring to a group of co-occurring repeats that differ by only a few nucleotides. Finally, Haft et al. (12) used REPfind (http://bibiserv.techfak.uni-bielefeld.de/reputer/), a part of the REPuter package (22–24) and BLASTN to identify smaller repeat clusters.
These programs are the most used tools in CRISPR detection, although none of them is especially conceived for this purpose. They require further manual manipulations to eliminate background data (tandem repeats for example) and importantly, do not define accurately the DR consensus (due to errors on the boundaries). Recently, two CRISPR- dedicated software tools were proposed, CRT (http://www.room220.com/crt) and PILER-CR (25). Both of them run fast and perform well in finding CRISPRs. However, CRT results in a considerable background since tandem repeats are considered as putative CRISPRs and in addition, the same CRISPR is sometimes detected more than once with different consensus DRs. PILER-CR has also some drawbacks since it often misidentifies the DR boundaries and omits the truncated DR.
In addition, there is no user-friendly dedicated web site. A specialized program to automatically identify CRISPRs seems to be mandatory for their optimum, rapid exploration and in-depth analysis, in order to increase the efficiency of CRISPRs investigations. CRISPRFinder is a web service offering fundamental tools for CRISPR detection, including the shortest ones, allowing an accurate definition of the DR consensus boundaries and extraction of the related spacers. It offers also additional tools to analyze the CRISPR loci: (i) obtain the CRISPR and the flanking sequences according to flexible size; (ii) make a blast of selected spacers or flanking sequences against the Genbank database and (iii) check if the DR is found elsewhere in prokaryotic sequenced genomes. The CRISPRFinder web interface is accessible through http://crispr.u-psud.fr/Server/CRISPRfinder.php
METHODS AND IMPLEMENTATION
CRISPRFinder core routines were developed in Perl under Debian Linux. The input of the web tool is a genomic query sequence of length up to 67 Mb in ‘FASTA’ format. Possible locations of CRISPRs (consisting of at least one motif) are detected by finding maximal repeats. A maximal repeat (26) is a repeat that cannot be extended in either direction without incurring a mismatch. The total number of maximal repeats in a sequence of size n is linear (less than n) which is interesting since the computation may be done in linear time using a suffix-tree-based algorithm. A CRISPR pattern of two DRs and a spacer may be considered as a maximal repeat where the repeated sequences are separated by a sequence of approximately the same length.
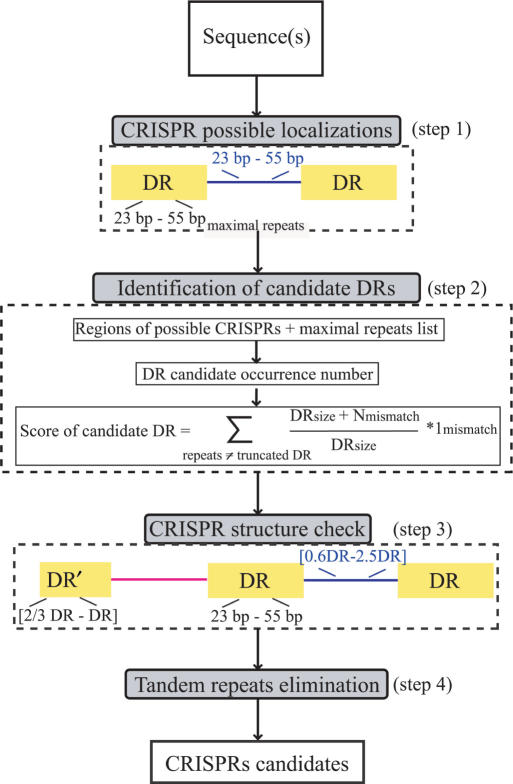
The operation of the program can be divided into four main steps summarized in Figure 1: (Step 1) browsing the maximal repeats of length 23–55 bp interspaced by sequences of 25–60 bp, (Step 2) selecting the DR consensus according to a defined score taking into account the number of occurrences of the candidate DR in the whole genome and privileging internal mismatches between the DRs rather than mismatches in the first or the last nucleotides, (Step 3) defining candidate CRISPRs after checking if they fit CRISPR definition, (Step 4) eliminating residual tandem repeats.
Figure 1.
CRISPR Finder flow chart. (Step 1) Browsing the maximal repeats to get possible CRISPR localizations using the Vmatch program. (Step 2) Consensus DR selection according to candidate occurrences and a score computation: the score privileges internal mismatches between direct repeats of a cluster rather than boundary mismatches. (Step 3) DR and spacers size check. (Step 4) Tandem repeats elimination using ClustalW for aligning spacers.
In the first step, maximal repeats are found by the software Vmatch (http://www.vmatch.de/), the upgrade of REPuter (22–24). Vmatch is based on a comprehensive implementation of enhanced suffix arrays (27) which provides the power of suffix trees with lower space requirements. A one nucleotide mismatch is allowed permitting minimal CRISPRs with a single nucleotide mutation between DRs to be found. Hereafter, the obtained maximal repeats are grouped to define regions of possible CRISPRs with a display of consensus DR candidates related to each cluster.
The second step is aimed at retrieving the DR consensus of each cluster. The difficulty resides especially in the identification of boundaries, which is very important to extract the correct spacers and compare DRs. In fact, the consensus DR is selected as the maximal repeat which occurs the most in the whole underlying genome sequence with respect to the forward and the reverse complement directions (since two CRISPRs having the same DR consensus may be in opposite directions). Thus, ambiguity in the choice of a DR will be eliminated in the case of presence of similar DRs in other CRISPRs of the related genomic sequence. However, if occurrence numbers are equal, more than a single DR consensus candidate are kept and later compared. Given a candidate consensus DR, the pattern search program fuzznuc of the EMBOSS package (28) is applied to get DRs’ positions in the related cluster. As the first or the last DR in a CRISPR may be diverged/truncated, a mismatch of one-third of the DR length is allowed between the flanking DRs and the candidate consensus DR, whereas smaller nucleotide differences are allowed between the other DRs to take into account possible single mutations. In case of multiple DR candidates, a score is computed and the best one (minimum) is picked. This score favours candidates which are encountered more frequently, rather than consensus DR showing less internal mismatches.
Once the DR consensus is determined, the corresponding spacers (Step 3) are extracted according to the DR boundaries determined previously. The spacer length is not allowed to be shorter than 0.6 or longer than 2.5 times the DR length. These sizes are in the range of CRISPRs described in the literature.
The last step consists in discarding false CRISPRs. Therefore, tandem repeats are eliminated by comparing the consensus DR with the spacer if there is only one spacer, or by comparing spacers between each other. The comparison is done with the CLUSTALW program (29) and the percentage of identity between spacers is not allowed to exceed 60%. Finally, candidates having at least three motifs and at least two exactly identical DRs are considered as confirmed CRISPRs. The remaining candidates are considered as questionable. These should be critically investigated by, for example, checking for intraspecies size variation of the locus.
INPUT AND OPTIONS
The query sequence must be in ‘FASTA’ format. Ns characters are accepted, IUB/GCG letters (MRWSYKVHDBX) will be converted to Ns and considered as mismatches but any other characters will be deleted. One can either paste the genomic sequence into the input field or upload it from a file on the local machine. Multisequence files are also allowed by the program and will be treated independently. Users may use the default version or click on the ‘advanced version’ link to set and modify all the program parameters, which may be especially useful for fixing the DR size.
OUTPUT
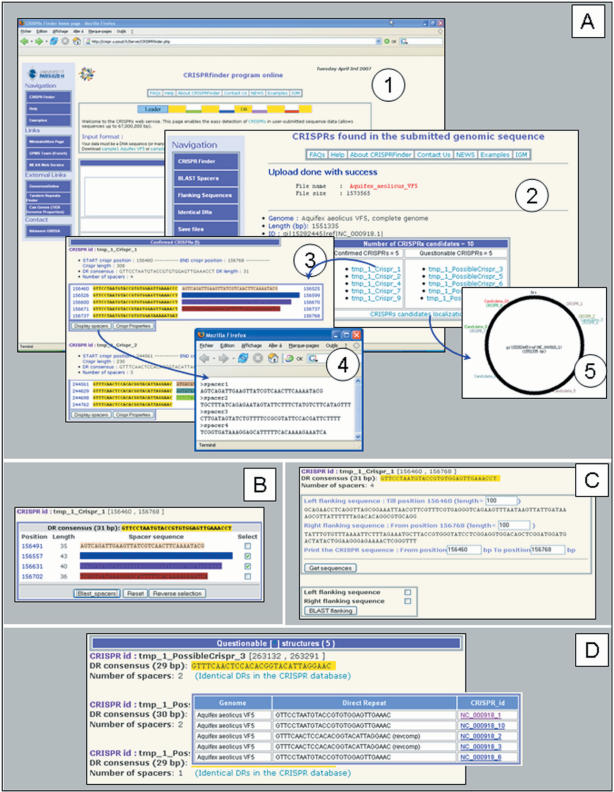
After querying a genomic sequence by CRISPRFinder, results are summarized in a table with the number of confirmed and questionable CRISPRs (Figure 2A). A CRISPR locus is presented according to a colour code showing DRs in yellow and spacers in different colours. The respective positions are displayed, in addition to links to two files: a summary of the displayed properties (number of motifs, DR consensus, positions, etc.) and a fasta file containing the list of spacers. In addition, a PNG (Portable Network Graphics) figure displays the different candidates’ location in the analysed sequence. In the case of presence of CRISPR clusters, further analysis may be done through three hyperlinks in the left menu: (i) blast spacers against the Genbank databases with a cutoff of 0.1 for the E-value and a matching length of at least 70% the queried spacer size (Figure 2B); (ii) obtain CRISPR and flanking sequences which are especially useful to define the leader sequence. As the size of the leader sequence depends on the species (it varies from 100 to 500 bp), the retrieved sequence may be manually modified by the user (Figure 2C) and (iii) display identical DRs in other known CRISPR loci (Figure 2D). This utility corresponds to a link to CRISPR database (Grissa et al. submitted for publication).
Figure 2.
An example of CRISPRFinder output using the Aquifex aeolicus VF5 genomic sequence (Refseq: NC_000918). (Panel A) (1) Home page where the genomic sequence is submitted. (2) Table listing the detected CRISPRs candidates (questionable and confirmed) providing links to each one. (3) CRISPRs details, the DR is showed in yellow and the spacers in different colours. (4) A fasta file displaying the first CRISPR spacers. (5) Figure showing the Aquifex circular chromosome with CRISPRs positions. (Panel B) One or several spacers may be blasted against NCBI databases by clicking on the blast_spacers button. (Panel C) The flanking and the CRISPR sequences may be viewed by clicking on the Get sequences button. The sequences boundaries may be modified by the user. (Panel D) The list of consensus DRs for all CRISPRs is shown with a link to identical DRs in the CRISPR database.
DISCUSSION AND CONCLUSIONS
CRISPRFinder is a program that allows the identification of structures with the principal characteristics of CRISPRs, the smaller being composed of a truncated or diverged DR, a spacer and a complete DR. In their analysis, Godde et al. (20) using Patscan had chosen to retain only CRISPRs with at least three exact repeats (eliminating CRISPRs constituted of a first truncated repeat plus two exact repeats) thus ignoring most CRISPRs containing less than three spacers. Similarly in the work by Durand et al.(21), the PYGRAM program is mostly efficient in visually displaying large CRISPRs. Such stringent criteria were appropriate in order to avoid ambiguities in early investigations which were essentially describing these new structures. However, it is now important, in order to better understand the evolution and spreading of CRISPRs, to provide tools which will not eliminate the smallest CRISPRs. This is what we chose to achieve with CRISPFinder. The major drawback is that when looking for the shortest structures, such as those with a unique spacer, it is clear that the background of spurious candidates can be very high. The output of Patscan and CRT also contains a large quantity of noised data that needs a manual treatment.
CRISPRFinder is accessible on the web and submission is very simple. We provide several samples on the website as demonstrators. Upon submission of the complete genome of Aquifex aeolicus VF5 (sample1), five confirmed and five possible CRISPRs are displayed in the following pages. On the contrary, while using the webservice for Patscan (http://www-unix.mcs.anl.gov/compbio/PatScan/), it is necessary to first define a pattern (which is not straightforward) and it is not possible to seek for CRISPRs in a single genomic sequence but rather in an entire predefined database. In addition, Patscan requires a Sun machine for local implementation. Similarly, PYGRAM only runs on linux systems and its installation requires advanced skills. CRT requires either to install JRE (Java Runtime Environment) or compile the source files, and PILER-CR needs to be compiled before use. A comparison between layouts of available online programs (REPuter, Patscan, TRF) and of CRISPRFinder is provided in the Supplementary Data.
To check that CRISPRFinder was efficient in recovering all the CRISPRs from a genome, we compared the results to other available studies on CRISPRs (15,20). The data were generally in good agreement, the differences being always in the DR boundaries’ identification (more accurate with CRISPRFinder) or in the number of motifs found, as the truncated DR is sometimes neglected or short clusters are not detected with other programs. Interestingly, some strains were claimed to be devoid of CRISPRs by Godde and colleagues but proved to have short CRISPRs with CRISPRFinder, such as in different Shigella sp. (S. sonnei Ss046, S. flexneri 2a str. 301, S. flexneri 2a str), or even long CRISPRs such as in Pseudomonas aeruginosa UCBPP-PA14. The latter example is shown on the CRISPRfinder website (sample2), and as can be seen by using the BLAST spacer function, six spacers out of thirty six at two different CRISPR loci, correspond to a bacteriophage sequence (bacteriophages F116, B3, D3112, DMS3 and phi CTX).
The tools developed here will assist in future CRISPRs’ analysis. Furthermore, the possibility to identify CRISPRs containing one or two motifs may help understand how new CRISPRs are created. The very small candidates will need to be typed across different isolates within the same species or very closely related species to search for variations. For instance, as shown with the sample file provided on the website (YP1 Yersinia), five Yersinia pestis strains possess at the same CRISPR locus two to eight spacers, some being unique and others shared by two or more strains (10). This strain-dependent polymorphism is especially interesting for epidemiological and phylogenetic studies (30,31). A tool to easily create a dictionary of spacers from different strains is proposed in a CRISPR-dedicated web database (http://crispr.u-psud.fr/crispr/).
The CRISPRFinder web server is an interface to extract with precision and to further analyse CRISPRs from genomic sequences. Four main advantages may be cited: (i) short CRISPR-like structures are detected, they are labelled questionable but may be of great interest if later confirmed; (ii) DRs are accurately defined to single base pair resolution; (iii) summary files may be uploaded (CRISPR properties summary and spacers file in Fasta format) and (iv) flanking sequences or spacers can be easily extracted and blasted against different databases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The CNRS and Université Paris Sud 11 have funded this project. Work on the development of genotyping tools for bacterial pathogens is supported by the Délégation Générale pour l’Armement (DGA). Funding to pay the Open Access publication charges for this article was provided by ADGE.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mojica FJ, Ferrer C, Juez G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 1995;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 3.Mojica FJ, Diez-Villasenor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 4.Peng X, Brugger K, Shen B, Chen L, She Q, Garrett RA. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J. Bacteriol. 2003;185:2410–2417. doi: 10.1128/JB.185.8.2410-2417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Embden JD, van Gorkom T, Kremer K, Jansen R, van Der Zeijst BA, Schouls LM. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J. Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jansen R, van Embden JD, Gaastra W, Schouls LM. Identification of a novel family of sequence repeats among prokaryotes. OMICS. 2002;6:23–33. doi: 10.1089/15362310252780816. [DOI] [PubMed] [Google Scholar]
- 8.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 9.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 10.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 11.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 12.Haft DH, Selengut J, Mongodin EF, Nelson KE. A Guild of 45 CRISPR-Associated (Cas) Protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl Acad. Sci. U.S.A. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lillestol R, Redder P, Garrett R, Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 17.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mokrousov I, Narvskaya O, Limeschenko E, Vyazovaya A. Efficient discrimination within a Corynebacterium diphtheriae epidemic clonal group by a novel macroarray-based method. J. Clin. Microbiol. 2005;43:1662–1668. doi: 10.1128/JCM.43.4.1662-1668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 20.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 21.Durand P, Mahe F, Valin AS, Nicolas J. Browsing repeats in genomes: Pygram and an application to non-coding region analysis. BMC Bioinformatics. 2006;7:477. doi: 10.1186/1471-2105-7-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurtz S, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. Computation and visualization of degenerate repeats in complete genomes. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:228–238. [PubMed] [Google Scholar]
- 24.Kurtz S, Schleiermacher C. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics (Oxford, England) 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gusfield D. Algorithms on Strings, Tree and Sequences. NY: Cambridge University Press; 1997. [Google Scholar]
- 27.Abouelhoda M, Kurtz S, Ohlebusch E. Replacing suffix trees with enhanced suffix arrays. Journal of Discrete Algorithms. 2004;2:53–86. [Google Scholar]
- 28.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoe N, Nakashima K, Grigsby D, Pan X, Dou SJ, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, et al. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg. Infect. Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]