Abstract
‘FastContact’ is a server that estimates the direct electrostatic and desolvation interaction free energy between two proteins in units of kcal/mol. Users submit two proteins in PDB format, and the output is emailed back to the user in three files: one output file, and the two processed proteins. Besides the electrostatic and desolvation free energy, the server reports residue contact free energies that rapidly highlight the hotspots of the interaction and evaluates the van der Waals interaction using CHARMm. Response time is ∼1 min. The server has been successfully tested and validated, scoring refined complex structures and blind sets of docking decoys, as well as proven useful predicting protein interactions. ‘FastContact’ offers unique capabilities from biophysical insights to scoring and identifying important contacts.
INTRODUCTION
The most intuitive decomposition of the binding free energy involves four terms (1–3): van der Waals (vdW) interactions, electrostatic, hydrophobicity and configurational entropy. The relative contribution of the changes between the bound and free states of these four terms is not the same. For stability (1), the main contributions appear to be electrostatic and desolvation interactions. For refined docked conformations, vdW interactions are expected to balance between the bound and unbound state, as they seemingly do in protein folding (1). This is good news, since it is not yet possible to readily estimate solute–solvent interactions. It should be noted, however, that solute–solute vdW has been shown to be an important consideration for complex refinement (4). Configurational entropy loss upon binding, including rotational and translational degrees of freedom, is always important, rough estimates based on crystal complexes varying between 5 and 15 kcal/mol (2,5–8). For the most part, this entropy depends on the flexibility of the unbound or free state with respect to the bound, with smaller corrections depending on the docking geometry. Since there is no robust estimate of entropy for a given protein, empirical free energy estimates, like ‘FastContact’, are always subject to an entropic correction. Hence, the server is most useful for discrimination between protein–protein docked complexes, and, more generally, for identifying energetically important contacts at the interface.
‘FastContact’, originally published in (9,10) rapidly estimates the electrostatic and desolvation component of the free energy based on a classic distance dependent dielectric 4r (11) and an empirical contact potential for the desolvation contribution (7) developed using a database of crystal (no complexes) structures from the PDB. Because of the pairwise nature of the empirical interactions, ‘FastContact’ is also able to report the contribution of individual residues and pairs of residues to the free energy. The latter should prove useful for site-directed mutagenesis studies since rankings of these interactions consistently identify the hot spots in the interface.
BRIEF DESCRIPTION OF THE ALGORITHM
The code behind the server was written in Fortran 77 and the server itself was written in PHP. ‘FastContact’ performs a fast computational estimate of the binding free energy between two proteins based on atomic pairwise interactions:
Electrostatic energy: the standard intermolecular Coulomb electrostatic potential with a distant-dependent dielectric constant equal to 4r, enforcing a minimum atom-to-atom distance separation equal to the sum of their corresponding vdW radii to avoid artificial overlaps.
Desolvation free energy: knowledge-based contact potential that accounts for hypdophobic interactions, self-energy change upon desolvation of charged and polar atom groups and side-chain entropy loss.
vdW energy: the standard 6–12 Lennard–Jones potential is evaluated using the program CHARMm (12) as part of the optimization of polar Hydrogens and overlaps.
The first two values (i–ii) can be used to calculate the overall free energy of the protein–protein interactions, assuming solute and/or solvent vdW cancellation between the bound and free proteins, and a correction factor for the configurational entropy loss. The application uses the definition of the atomic composition of each amino acid consistent with CHARMm19 parameters.
USING THE FASTCONTACT SERVER
Required user-input information
Figure 1 shows a snapshot of the input page. The user uploads two Protein Data Bank (PDB) format files (13), one ‘receptor’ and one ‘ligand’, along with their email address. The web server currently makes no distinction between chains; it simply reads in each line in the PDB file starting with an ‘ATOM’ field. The maximum number of residues is limited to 1500. The email address is where the output/results will be sent (as a file attachment). Hydrogen bonds and missing atoms are built and optimized on the uploaded structures using the molecular software CHARMm.
Figure 1.
Snapshot of input page.
Optional parameters
Range of desolvation interaction
The default range is 6 Å, such that the potential smoothly goes to zero between 5 and 7 Å. This range is suggested for refined models, without overlaps and relatively snuggly fit interfaces, e.g. (4,10). The user has the option of changing the range to 9 Å, approaching zero between 8 and 10 Å. This modality is suggested for encounter complexes, e.g. (14,15) for a rigid body docking validation.
Minimization
The default setting for Hydrogen bond optimization and removal of minimal overlaps prescribes a short 3 × 20 ABNR minimization steps with fixed backbone using the program CHARMm and the PARAM19 residue topology file (RTF). However, the user is free to change this setting to a full atom minimization. This setting will work for single chains only and no gaps.
Patch end terminals with –NH3 + and –COOH
By default, the end terminal residues will be patched by CHARMm. In case the end terminals are missing from the structure, the user has the option of turning the patching feature off.
Output format and explanation
The results from a ‘FastContact’ server run are returned to the user via email as a file attachment (with a normal response time of ∼1 min). The attached file is a gzipped archive (.tar.gz) containing three results files: (i) the main results file (‘output.txt’); and, the processed (including H-bonds) and renumbered (ii) receptor PDB file (‘protein1’) and (iii) the ligand PDB file (‘protein2’). All of the files are prefixed with the user name (email prefix) and timestamp of the server run for easy reference.
The main source of errors in the output file relates with the format of the input PDB files. For instance, columns usage must strictly follow the PDB standards, and ATOM keyword must describe only protein amino acids. The server cannot minimize the backbone of sequences with gaps, and missing heavy atoms are sometimes not able to be reconstructed by the server. If the server detects an error, it will report a message with possible problems and suggestions.
The main results file (‘output.txt’) returns two components of a free energy function, electrostatic energy and desolvation free energy, and evaluates the solute vdW energy using CHARMm. The latter is sometimes useful to compare between different models (16), but here it is given only as a reference since it is not used in the analysis of contacts. Often vdW energies larger than about −500 kcal/mol suggest structural overlaps. Although ‘FastContact’ smoothes the potentials to tolerate some limited overlaps, these are, in general, detrimental to the quality of the computational estimates. Figure 2 shows the summary energy output and part of the contact analysis, for the barnase–barstar complex 1BRS. We should caution that, when submitting co-crystallized receptor and ligand structures, the automated minimization implemented in the server leads to an over optimization of the electrostatic contacts of ∼10–20% (5). The reason is because the direct electrostatic term used in the server does not have an angular dependence for Hydrogen bonds. Hence, these interactions tend to ‘double-dip’. This effect is compensated when scoring unbound models that always have some built in frustration due to the less optimal backbone and side chain conformations.
Figure 2.
Output page. It includes a summary of electrostatics, desolvation and vdW energies, followed by a list of the 20 most attractive and 20 most repulsive residues and contacts for the electrostatic, desolvation, and the sum of these two components that correlates with the binding free energy. For modeling, the repulsive information is sometimes useful as an indication of a wrong structural motif.
DISCUSSION
Critical assessment of protein interactions (CAPRI)
The method implemented in ‘FastContact’ has been successfully applied in the CAPRI experiment both as a free energy filtering procedure of the ‘ClusPro’ server (14) that predicts protein complexes and in protein–protein refinement (4) (using a 9 and 6 Å desolvation range, respectively). ‘FastContact’ has been instrumental in the success of our group in blind predictions (17,18). In rounds 1 and 2 of CAPRI, Camacho and Gatchell (19) produced some of the best model structures, appropriately distinguishing between near-native and false positive structures for three targets. In rounds 3–5, the automated server ‘ClusPro’ (the only server participating in CAPRI) predicted good models for 5 targets (15), while our manual predictions resulted in good predictions for 6 targets (20) (missing the 3 targets that had a significant structural rearrangement upon binding).
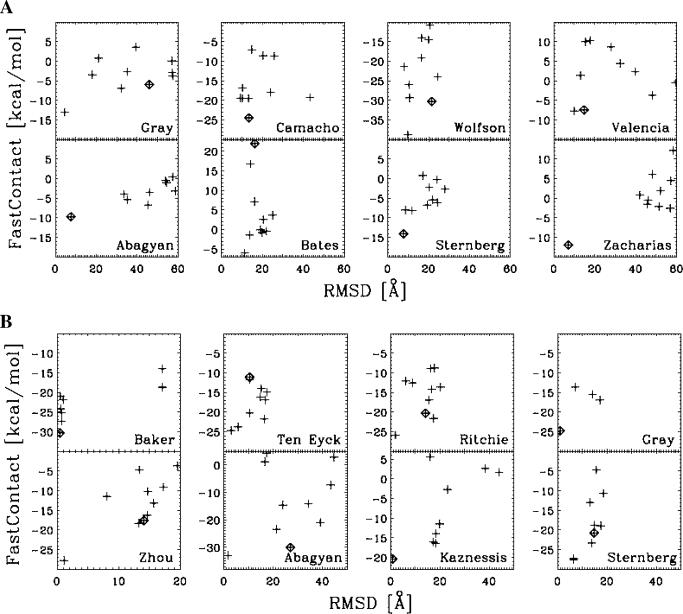
The robustness of our method was further supported by the analysis of the full set of models submitted for CAPRI (rounds 3–5) for the 6 targets that did not undergo a large structural rearrangement upon binding (18). For these targets, we showed that ‘FastContact’ was able to discriminate near-native predictions from docked conformations far from the binding site for 5 of the targets (10), and for all but one of the manual predictions submitted to CAPRI. For instance, Figure 3 shows the re-scoring of models submitted for targets 8 and 12 by 13 different groups around the world. In all cases, ‘FastContact’ correctly identified the near-native conformation, even when the modeler failed to do so.
Figure 3.
Examples of ‘FastContact’ scoring for a subset of high quality docked models from eight groups for targets 8 and 12 of CAPRI rounds 3–5, from http://capri.ebi.ac.uk/. For each of these targets, we run the models in our server and re-rank the models accordingly. In all cases, the server was able to correctly rank a low RMSD model as the one with the lowest free energy score. For comparison, we also marked with a diamond symbol the model ranked number 1 by the modeler (23–33). (A) target 8; (B) target 12. For a complete analysis of all models please see reference (10).
By splitting the free energy between electrostatics and desolvation, ‘Fastcontact’ also provides immediate insights into the nature of the binding interactions. Namely, negative desolvation is associated with a hydrophobic pocket at the binding site, whereas positive desolvation characterizes mostly polar interfaces. This is important since sometimes electrostatic or desolvation alone could lead to better discrimination than the combination of the two (21,22). The latter is, of course, due to the intrinsic limitations of empirical free energies. In particular, reliable estimates for solvent and entropic interactions are not yet available.
OTHER SERVERS
We are aware of only one server that estimate binding free energies of complex structures: http://sparks.informatics.iupui.edu/czhang/complex.html by the Zhou Lab (8). The server returns a single binding free energy estimate in kcal/mol that was shown to correlate with experimental values (±1.8 kcal/mol) for some 69 crystal structures.
AVAILABILITY
The web server is available freely and without registration at: http://structure.pitt.edu/servers/fastcontact/
ACKNOWLEDGEMENTS
The ‘FastContact’ Server has been thoroughly tested by over 500 runs from users all over the world. We are grateful to the many people around the world who tested our server and provided constructive feedback. This material is based upon work supported by the National Science Foundation under Grant No. MCB-0444291. Funding to pay the Open Access publication charges for this article was provided by NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bueno M, Camacho CJ, Sancho J. SIMPLE estimate of the free energy change due to aliphatic mutations: superior predictions based on first principles. Proteins. 2007 doi: 10.1002/prot.21453. (in press) [DOI] [PubMed] [Google Scholar]
- 2.Novotny J, Bruccoleri RE, Saul FA. On the attribution of binding energy in antigen-antibody complexes McPC 603, D1.3, and HyHEL-5. Biochemistry. 1989;28:4735–4749. doi: 10.1021/bi00437a034. [DOI] [PubMed] [Google Scholar]
- 3.Vajda S, Weng Z, DeLisi C. Extracting hydrophobicity parameters from solute partition and protein mutation/unfolding experiments. Protein Eng. 1995;8:1081–1092. doi: 10.1093/protein/8.11.1081. [DOI] [PubMed] [Google Scholar]
- 4.Camacho CJ, Vajda S. Protein docking along smooth association pathways. Proc. Natl Acad. Sci. USA. 2001;98:10636–10641. doi: 10.1073/pnas.181147798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura SR, Brower RC, Vajda S, Camacho CJ. Dynamical view of the positions of key side chains in protein-protein recognition. Biophys. J. 2001;80:635–642. doi: 10.1016/S0006-3495(01)76044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vajda S, Weng Z, Rosenfeld R, DeLisi C. Effect of conformational flexibility and solvation on receptor-ligand binding free energies. Biochemistry. 1994;33:13977–13988. doi: 10.1021/bi00251a004. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Vasmatzis G, Cornette JL, DeLisi C. Determination of atomic desolvation energies from the structures of crystallized proteins. J. Mol. Biol. 1997;267:707–726. doi: 10.1006/jmbi.1996.0859. [DOI] [PubMed] [Google Scholar]
- 8.Liu S, Zhang C, Zhou H, Zhou Y. A physical reference state unifies the structure-derived potential of mean force for protein folding and binding. Proteins. 2004;56:93–101. doi: 10.1002/prot.20019. [DOI] [PubMed] [Google Scholar]
- 9.Camacho CJ, Zhang C. FastContact: rapid estimate of contact and binding free energies. Bioinformatics. 2005;21:2534–2536. doi: 10.1093/bioinformatics/bti322. [DOI] [PubMed] [Google Scholar]
- 10.Camacho CJ, Ma H, Champ PC. Scoring a diverse set of high-quality docked conformations: a metascore based on electrostatic and desolvation interactions. Proteins. 2006;63:868–877. doi: 10.1002/prot.20932. [DOI] [PubMed] [Google Scholar]
- 11.Pickersgill RW. A rapid method of calculating charge-charge interaction energies in proteins. Protein Eng. 1988;2:247–248. doi: 10.1093/protein/2.3.247. [DOI] [PubMed] [Google Scholar]
- 12.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 13.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comeau SR, Vajda S, Camacho CJ. Performance of the first protein docking server ClusPro in CAPRI rounds 3-5. Proteins. 2005;60:239–244. doi: 10.1002/prot.20564. [DOI] [PubMed] [Google Scholar]
- 16.Camacho CJ, Gatchell DW, Kimura SR, Vajda S. Scoring docked conformations generated by rigid-body protein-protein docking. Proteins. 2000;40:525–537. doi: 10.1002/1097-0134(20000815)40:3<525::aid-prot190>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 17.Mendez R, Leplae R, De ML, Wodak SJ. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 18.Mendez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3-5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 19.Camacho CJ, Gatchell DW. Successful discrimination of protein interactions. Proteins. 2003;52:92–97. doi: 10.1002/prot.10394. [DOI] [PubMed] [Google Scholar]
- 20.Camacho CJ. Modeling side-chains using molecular dynamics improve recognition of binding region in CAPRI targets. Proteins. 2005;60:245–251. doi: 10.1002/prot.20565. [DOI] [PubMed] [Google Scholar]
- 21.Camacho CJ, Weng Z, Vajda S, DeLisi C. Free energy landscapes of encounter complexes in protein-protein association. Biophys. J. 1999;76:1166–1178. doi: 10.1016/S0006-3495(99)77281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 23.Tress M, de JD, Grana O, Gomez MJ, Gomez-Puertas P, Gonzalez JM, Lopez G, Valencia A. Scoring docking models with evolutionary information. Proteins. 2005;60:275–280. doi: 10.1002/prot.20570. [DOI] [PubMed] [Google Scholar]
- 24.Mustard D, Ritchie DW. Docking essential dynamics eigenstructures. Proteins. 2005;60:269–274. doi: 10.1002/prot.20569. [DOI] [PubMed] [Google Scholar]
- 25.Carter P, Lesk VI, Islam SA, Sternberg MJ. Protein-protein docking using 3D-Dock in rounds 3, 4, and 5 of CAPRI. Proteins. 2005;60:281–288. doi: 10.1002/prot.20571. [DOI] [PubMed] [Google Scholar]
- 26.Zacharias M. ATTRACT: protein-protein docking in CAPRI using a reduced protein model. Proteins. 2005;60:252–256. doi: 10.1002/prot.20566. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez-Recio J, Abagyan R, Totrov M. Improving CAPRI predictions: optimized desolvation for rigid-body docking. Proteins. 2005;60:308–313. doi: 10.1002/prot.20575. [DOI] [PubMed] [Google Scholar]
- 28.Daily MD, Masica D, Sivasubramanian A, Somarouthu S, Gray JJ. CAPRI rounds 3-5 reveal promising successes and future challenges for RosettaDock. Proteins. 2005;60:181–186. doi: 10.1002/prot.20555. [DOI] [PubMed] [Google Scholar]
- 29.Smith GR, Fitzjohn PW, Page CS, Bates PA. Incorporation of flexibility into rigid-body docking: applications in rounds 3-5 of CAPRI. Proteins. 2005;60:263–268. doi: 10.1002/prot.20568. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, Liu S, Zhou Y. Docking prediction using biological information, ZDOCK sampling technique, and clustering guided by the DFIRE statistical energy function. Proteins. 2005;60:314–318. doi: 10.1002/prot.20576. [DOI] [PubMed] [Google Scholar]
- 31.Inbar Y, Schneidman-Duhovny D, Halperin I, Oron A, Nussinov R, Wolfson HJ. Approaching the CAPRI challenge with an efficient geometry-based docking. Proteins. 2005;60:217–223. doi: 10.1002/prot.20561. [DOI] [PubMed] [Google Scholar]
- 32.Schueler-Furman O, Wang C, Baker D. Progress in protein-protein docking: atomic resolution predictions in the CAPRI experiment using RosettaDock with an improved treatment of side-chain flexibility. Proteins. 2005;60:187–194. doi: 10.1002/prot.20556. [DOI] [PubMed] [Google Scholar]
- 33.Law D, Hotchko M, Ten EL. Progress in computation and amide hydrogen exchange for prediction of protein-protein complexes. Proteins. 2005;60:302–307. doi: 10.1002/prot.20574. [DOI] [PubMed] [Google Scholar]