Abstract
We designed a web server for the analysis of biosynthetic capacities of metabolic networks. The implementation is based on the network expansion algorithm and the concept of scopes. For a given network and predefined external resources, called the seed metabolites, the scope is defined as the set of products which the network is in principle able to produce. Through the web interface the user can select a variety of metabolic networks or provide his or her own list of reactions. The information on the organism-specific networks has been extracted from the KEGG database. By choosing an arbitrary set of seed compounds, the user can obtain the corresponding scopes. With our web server application we provide an easy to use interface to perform a variety of structural and functional network analyses. Problems that can be addressed using the web server include the calculation of synthesizing capacities, the visualization of synthesis pathways, functional analysis of mutant networks or comparative analysis of related species. The web server is accessible through http://scopes.biologie.hu-berlin.de.
INTRODUCTION
The ever increasing number of fully sequenced genomes results in a rapidly expanding knowledge of the metabolic capabilities of a wide variety of organisms. This information is collected and made accessible through biochemical databases such as KEGG (1), Brenda (2) or BioCyc (3).
Whereas our knowledge on the wiring of the metabolic networks is far advanced, precise data on the kinetic properties of the catalyzing enzymes is still sparse. However, even without the ability to formulate kinetic models, the topology alone can be used for a wide variety of structural analyses, from which informative properties such as principle biosynthetic capabilities or feasible flux distributions can be derived. Established structural approaches include the concept of elementary flux modes (4,5), the closely related concept of extreme fluxes (6), flux balance analysis (7) as well as graph theoretical approaches (8,9).
We have recently introduced the concept of network expansion for the structural analysis of large-scale metabolic networks (10). The algorithm allows to calculate for a given network and predefined external resources (the seed compounds) those chemical compounds which the network is in principle able to produce. This set of products is called the scope of the seed. Because scopes characterize the synthesizing capacities of metabolic networks, this concept is well suited for relating structural to functional properties of the networks. With this method we explored the hierarchical structuring of metabolic networks, where we focused on a complete network comprising enzymatic reactions originating from a wide variety of organisms (11). We also compared metabolic capabilities of organism-specific networks (12) and developed a model of metabolic evolution (13). Furthermore, we analysed the changes of metabolic capacities in response to environmental perturbations (14).
These results demonstrate the general usefulness and wide applicability of the concept of network expansion. With the development of the here-described web server application ‘MetaPath Online’, we provide a public access to this method enabling scientists to investigate specific metabolic hypothesis on particular networks. Such hypotheses include the question whether certain metabolites can be produced by a particular organism and, if so, what may be a possible synthesis route. Moreover, with the inclusion of user specified sets, it is possible to analyse the metabolic performance of mutants in which one or several reactions are removed or added.
EXPANDING METABOLIC NETWORKS
Concept of scopes
The method of expanding networks can be described as follows: starting with given metabolites, called seed compounds, the algorithm selects from a database in an iterative manner those reactions, whose substrates are either part of the set of seed compounds or are products of reactions which were already selected in an earlier iteration step. This network expansion process ends when no further reactions fulfilling this condition can be found. All metabolites which can be produced by the resulting set of reactions form the scope of the seed compounds. Scopes describe therefore the synthesizing capacity of the corresponding seed compounds in a specified metabolic network.
In principle, enzyme catalysed reactions can proceed in forward as well as in backward direction. However, under physiological conditions the metabolite concentrations may be such that one direction is thermodynamically unfeasible. The expansion algorithm can consider both irreversible and reversible reactions, where reversible reactions are incorporated into the expanding network if either all their substrates or all their products are already present.
The size of a scope strongly depends on the specific choice of seed compounds as well as on the investigated metabolic network. Besides the scope, which is the final result of the expansion algorithm, the analysis of the expansion process itself is of interest. The expansion curve shows in many cases characteristic features which depend on structural properties of the network (10).
Impact of cofactors
In metabolic networks, many reactions require the presence of particular metabolites, so-called cofactors, which typically participate in a large number of reactions and are responsible for specific functions. For example, in the reaction catalysed by the enzyme hexokinase,
| 1 |
ATP acts as a cofactor by transferring one phosphate group to glucose. This pattern occurs in a large number of reactions and can be considered as a main function of ATP. Due to its importance, a cell in a typical physiological state ensures that ATP is always present in a sufficient amount. When asking for products which can be synthesized from glucose in such a cell, it is therefore reasonable to assume that a reaction such as (1) can proceed.
To account for this, we have implemented a variant of the expansion algorithm in which reactions utilizing ATP as a cofactor can also be included even if ATP has not been synthesized from the seed compounds. The algorithm ensures that in such a case ATP can only be used in its function as cofactor, i.e. as phosphate donor, but not as a substrate for the synthesis of other metabolites.
In a similar way, we implemented other variants accounting for the presence of the important cofactors NADH/NAD+ and NADPH/NADP+ which mediate redox reactions, as well as CoA which is a carrier of acyl groups.
The consideration of cofactors may drastically influence the expansion process. In general, the expansion in presence of a cofactor leads to a larger scope than the corresponding process without the cofactor. Furthermore, the velocity of the expansion process may be increased considerably.
Extraction of synthesis pathways
A scope includes those metabolites which can be synthesized from a specified set of seed compounds. An important biochemical question is how a particular product can be synthesized with a minimal number of reactions from a given set of substrates.
We have designed an algorithm which extracts from an expanded network a set of reactions allowing for the synthesis of the desired product (the target) in a minimal number of consecutive steps. The algorithm works as follows: starting from the seed, an expansion process is performed as described earlier. The number of consecutive synthesis steps necessary to produce the target is already determined by the iteration step in which the target was incorporated. During the process, it is recorded which metabolites enable which reactions and which reactions produce which metabolites. With this information, the synthesis pathway is now assembled in a reverse order starting from the target. It should be noted that this definition of a pathway somewhat differs from the usual textbook notion. The determined sets of reactions represent a synthesis route by which the target metabolite—and possibly some side products—can be produced while exclusively consuming the seed metabolites.
The obtained pathways in general are not simply unbranched chains but contain reactions which operate in parallel. There may exist other pathways with a smaller total number of reactions which can perform the same conversion, however none with a smaller number of consecutive steps.
WEB SERVER IMPLEMENTATION
We have developed a web-based tool which provides public access to the algorithms described earlier. The user may select for his or her analysis a wide range of metabolic networks, including organism-specific metabolic networks from a list of over 400 species as well as a reference network comprising over 5000 reported reactions or define a customized network by uploading a list of reactions. The user may choose to include information on the reversibility of reactions or to consider all reactions as reversible. The latter choice may be reasonable because for some reactions the information on reversibility is arguable, in particular since the direction of a reaction may depend on environmental conditions or the specific cell type.
In its current state, the web server offers three types of analyses. It is possible to calculate the scopes of arbitrary sets of seed compounds, visualize the course of the expansion process and identify shortest synthesis pathways between chemical compounds. These three applications are described in more detail later in this chapter.
Data import
The information on metabolic networks has been extracted from the KEGG database. From the LIGAND subdivision (plain text file as of 13 February 2007), the complete list of 6825 reactions has been imported. The reactions have been checked for consistency. We rejected 290 reactions because they showed an erroneous stoichiometry, by which we mean that some atomic species occurred in different numbers on both sides of the reaction. The inclusion of such erroneous reactions could result in absurd events such as the creation of chemical elements or groups. We identified compounds possessing ambiguous structure information, such as chains of chemical groups of unspecified length (e.g. Ubiquinol, KEGGID: C00390, C14H20O4[C5H8]n) or compounds with unspecified residues (e.g. Amino acid, KEGGID: C00045, C2H4NO2R). We rejected 990 reactions involving such metabolites. Furthermore, we did not include 342 reactions involved in glycan synthesis because the focus of our application lies on the metabolism of small chemical species which also does not include the formation of complex structures such as proteins or RNA and DNA molecules. Of the remaining reactions, four display identical stoichiometries, leaving 5199 unique reactions which are used as the data set for the web server application.
Furthermore, information on the reversibility of reactions has been extracted from the KGML files which specify the pathways for all organisms included in KEGG. In general, a particular reaction is listed in several KGML files and the information on its reversibility may be ambiguous. In fact, we identified 136 reactions for which this is the case. For our web server application, we consider a reaction to be irreversible only if it is defined as irreversible in all corresponding occurrences in the KGML files. This is the case for 2622 of the 5199 reactions. For the calculations, the user can decide whether he wishes to include this information on reversibility or not.
The organism-specific networks were determined using the files ‘reaction’ and ‘enzyme’ contained in the KEGG/LIGAND database. In the first step, for all reactions the EC numbers of the catalyzing enzymes were retrieved from their corresponding entries in the ‘reaction’ file (section ENZYME). Subsequently, from the ‘enzyme’ file, for each enzyme a list of organisms is obtained in which there exists a corresponding gene (section GENES). Thus, for each organism the metabolic network is defined by all those reactions for which a catalyzing enzyme is encoded in its genome. In all cases where an enzyme is not fully classified (e.g. EC1.3.1.-), the corresponding entry in the file ‘enzyme’ does not contain a section GENES. As a consequence, no such reactions are included in organism-specific networks. This also implies that the union of all organism-specific networks is significantly smaller (2589 reactions) than the reference network. This may result in drastically different scopes when comparing the reference network with the union of all organism networks.
Scope calculation
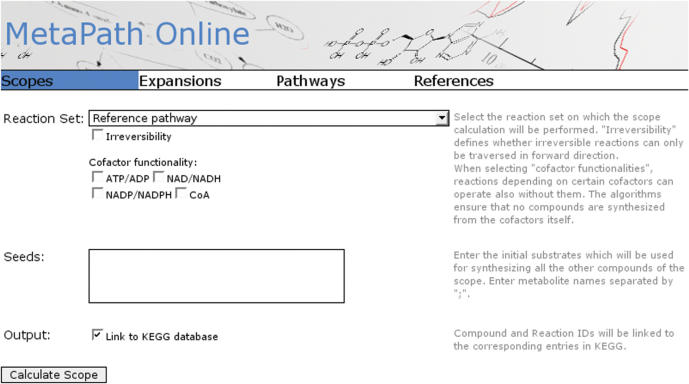
Figure 1 depicts the initial screen of the web application. The preselected function is the calculation of scopes. The network to be investigated can be selected from a drop-down menu. The first check box is used to select whether information on reversibility should be included, the other four boxes allow to select which cofactors should be considered to be present. In the text field the user enters a list of seed compounds separated by semicolons. With the check box at the bottom of the screen the user can select whether the resulting compounds and reactions appear as hyperlinks to their corresponding entries on the KEGG website.
Figure 1.
The initial screen of MetaPath Online containing the input mask for the scope calculation.
The choice of available networks includes the reference network (comprising all 5199 reactions), 488 organism-specific networks, the union of these organism-specific networks, as well as the option to enter a user defined network. In case the latter option is selected, a further text field appears in which a comma-separated list of KEGG reaction identifiers can be entered. The identifiers can be prepended by ‘+’ or ‘–’ to indicate whether the corresponding reaction is irreversible in the forward or backward direction (as defined in KEGG), respectively. It should be noted that the same consistency tests that are applied to the original KEGG reaction sets are also applied to the user-specified networks.
The seed compounds may be specified as KEGG identifiers or by common names. In the latter case, the application tries to match the input with the compound names specified in the KEGG/LIGAND database. The matching is case insensitive and ignores special characters. For example, ‘dglucose’, ‘d-Glucose’ and ‘d glucose’ are all matched to KEGG entry C00031, while ‘glucose’ or ‘alpha-d-glucose’ specify different compounds and are accordingly matched to C00293 and C00267, respectively.
The result is presented as a list of the compounds of the scope. Additionally, the program indicates which of the seed compounds could be identified.
Visualization of the expansion process
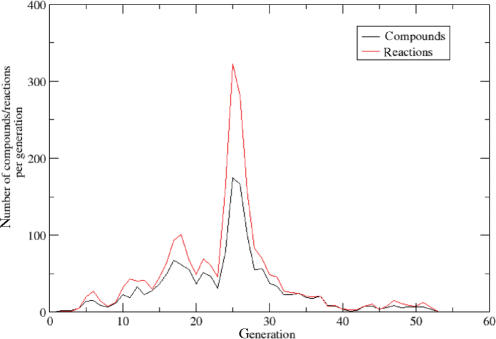
The input mask of this function has the same format as for the scope calculation. However, a different output is generated. First, a graphical representation of the expansion process is drawn (Figure 2). Secondly, two lists are produced containing the compounds and reactions within the expanded network, respectively. For each entry, also the number of the iteration step in which it was included into the network is given.
Figure 2.
Example expansion curve: shown is the expansion process on the reference network starting with ATP as seed.
Extraction of minimal synthesis pathways
For this function, the input mask is extended by a text field in which the target metabolite must be entered. The format is identical to that in the field for the seed compounds, however, only a single metabolite can be specified.
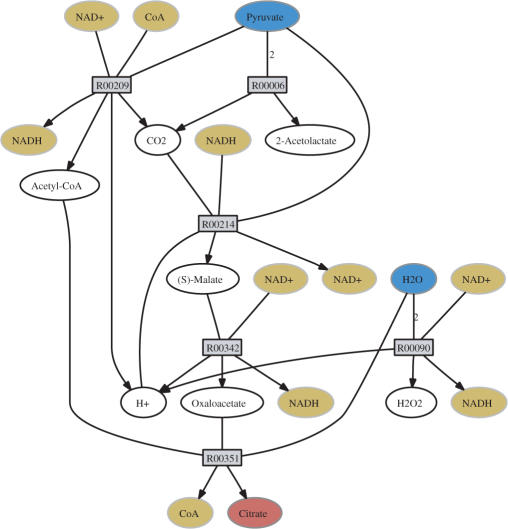
The output comprises the list of reactions occurring in the determined synthesis pathway. Furthermore, a graphical representation of this pathway is shown (for example, see Figure 3). For that, the calculated synthesis pathway is represented by a bipartite graph, with two groups of nodes representing the metabolites and reactions, respectively. Edges connect metabolites with the reactions they participate in. The edges point in that direction in which the corresponding reaction proceeds within the synthesis pathway. The final graph layout is performed by a hierarchical layout algorithm implemented in the program ‘dot’ from the publicly available graphviz package (15).
Figure 3.
Minimal synthesis path: the synthesis of citrate from pyruvate is shown. Irreversibility has been considered. The cofactors NAD+/NADH and CoA were available. The seed compounds (pyruvate, water and oxygen) are marked blue, the target metabolite (citrate) is marked red and the cofactors yellow.
Some metabolites may take part in a large number of reactions which would result in a confusing graphical layout. Therefore, it is convenient to draw compounds appearing in more than a certain number of reactions separately for each reaction they take part in. This threshold parameter can also be specified through the web interface.
DISCUSSION
The web server allows to analyse predefined metabolic networks which have been extracted from the KEGG database. In future, we plan to integrate other sources of biochemical information, by extracting networks from other databases such as BioCyc. The extension to a wider data source will increase the reliability of the results. However, metabolic databases are generally error-prone, and therefore curation of input data is an important prerequisite. In particular, the expansion process critically depends on correct stoichiometries because otherwise results would indicate that chemical species can be produced from nothing. Therefore, we applied very strict criteria for the acceptance of a reaction. On the other hand, due to this strictness, the algorithm might miss target metabolites which are in fact producible. To resolve this conflict, we plan to refine our curation procedure to allow more reactions, for example the inclusion of metabolites with chains of chemical groups of arbitrary length.
The possibility to analyse user-defined networks by uploading a list of KEGG reactions considerably extends the applicability of the tool because it lifts the restriction to predefined networks. In order to further widen the usability of our application, we intend to establish a function allowing the user to upload his or her own networks in a portable format, which may be the result of his or her own research activities. This makes the user independent from the reactions specified in the KEGG database allowing for the consideration of a wider spectrum of reactions.
CONCLUSIONS
In this work we presented ‘MetaPath Online’, a web server application for the structural and functional analysis of large-scale metabolic networks. It is based on the method of network expansion and can be used to calculate synthesizing capacities of over 400 species-specific networks as well as the reference network comprising all KEGG reactions. Specific scientific problems that can be addressed with this application include
Synthesis capacities: Can an organism under investigation produce particular metabolites if it is provided with specified resources?
Synthesis pathways: What is a possible reaction route to produce a given target metabolite from specified resources?
Mutant analysis: How do metabolic capabilities change under structural perturbations of the underlying network? In particular, how does the removal of one reaction or a complete pathway reduce the capacities and how does the extension of the network by a new pathway expand the capacities?
Comparative analysis: Are similarities of metabolic networks of related organisms reflected by similar metabolic capacities?
With the described implementation of our web server, we provide for a wide audience an easy to use interface for the network expansion method.
MetaPath Online is freely available for use at http://scopes.biologie.hu-berlin.de.
SUPPLEMENTARY DATA
Supplementary Data are available through the website.
ACKNOWLEDGEMENTS
The authors thank the German Research Foundation and the German Ministry of Education and Research for financial support. We thank Daniel Kahn for the inspiring discussions. Funding to pay the Open Access publication charges for this article was provided by the collaborative research center ‘Theoretical Biology’ (SFB 618) of the German Research Foundation.
Conflic of interest statment. None declared.
REFERENCES
- 1.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schomburg I, Chang A, Schomburg D. BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 2002;30:47–49. doi: 10.1093/nar/30.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuster S, Hilgetag C. On elementary flux modes in biochemical reaction systems at steady state. J. Biol. Syst. 1994;2:165–182. [Google Scholar]
- 5.Schuster S, Fell DA, Dandekar T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 2000;18:326–332. doi: 10.1038/73786. [DOI] [PubMed] [Google Scholar]
- 6.Papin JA, Price ND, Wiback SJ, Fell DA, Palsson BO. Metabolic pathways in the post)genome era. TIBS. 2003;28:250–258. doi: 10.1016/S0968-0004(03)00064-1. [DOI] [PubMed] [Google Scholar]
- 7.Kauffman KJ, Prakash P, Edwards JS. Advances in flux balance analysis. Curr. Opin. Biotechnol. 2003;14:491–496. doi: 10.1016/j.copbio.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Wagner A, Fell DA. The small world inside large metabolic networks. Proc. R. Soc. Lond. B. 2001;268:1803–1810. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 10.Handorf T, Ebenhöh O, Heinrich R. Expanding metabolic networks: scopes of compounds, robustness, and evolution. J. Mol. Evol. 2005;61:498–512. doi: 10.1007/s00239-005-0027-1. [DOI] [PubMed] [Google Scholar]
- 11.Handorf T, Ebenhöh O, Kahn D, Heinrich R. Hierarchy of metabolic compounds based on their synthesising capacities. IEE Proc. Syst. Biol. 2006;153:359–363. doi: 10.1049/ip-syb:20060021. [DOI] [PubMed] [Google Scholar]
- 12.Ebenhöh O, Handorf T, Heinrich R. A cross species comparison of metabolic network functions. Genome Informatics. 2005;16:203–213. [PubMed] [Google Scholar]
- 13.Ebenhöh O, Handorf T, Kahn D. Evolutionary changes of metabolic networks and their biosynthetic capacities. IEE Proc. Syst. Biol. 2006;153:354–358. doi: 10.1049/ip-syb:20060014. [DOI] [PubMed] [Google Scholar]
- 14.Ebenhöh O, Liebermeister W. Structural analysis of expressed metabolic subnetworks. Genome Informatics. 2006;17:163–172. [PubMed] [Google Scholar]
- 15.Gansner ER, Koutsofios E, North SC, Vo K-P. A technique for drawing directed graphs. IEEE Trans. Software Eng. 1993;19:214–230. [Google Scholar]