Abstract
KNOTS (http://knots.mit.edu) is a web server that detects knots in protein structures. Several protein structures have been reported to contain intricate knots. The physiological role of knots and their effect on folding and evolution is an area of active research. The user submits a PDB id or uploads a 3D protein structure in PDB or mmCIF format. The current implementation of the server uses the Alexander polynomial to detect knots. The results of the analysis that are presented to the user are the location of the knot in the structure, the type of the knot and an interactive visualization of the knot. The results can also be downloaded and viewed offline. The server also maintains a regularly updated list of known knots in protein structures.
INTRODUCTION
Interest in the topological properties of biological systems was greatly accelerated with the discovery of knots in single-stranded DNA in 1976 (1). Subsequently, knots in DNA were investigated extensively (2–5) and even created artificially in polymeric materials (6), but it took another 20 years before the first systematic studies of protein knots appeared (7–11). Topology is particularly relevant for proteins because the 3D structure of a protein directly determines its functionality. Recently, we performed a comprehensive analysis of the Protein Data Bank (11) and demonstrated that knotted structures tend to persist across species and kingdoms. However, when a knot appears or vanishes in the course of evolution, the function of the protein is also altered accordingly (11–13). We uncovered some knotted proteins that have significant biomedical importance, such as the Parkinson's disease-associated ubiquitin hydrolase UCH-L1 (14) or its structural homolog UCH-L3 (10,15), which contain the most complicated knots found in proteins so far. Other challenges include understanding the folding and unfolding of knotted proteins. The underlying mechanisms are not yet well understood and are the subject of active research (16,17).
Surprisingly, most discovered knots were not reported at the time the structure was solved, since finding knots in protein structures by naked eye is virtually impossible. Moreover, widely used protein structure verification tools like WHATIF (18), VERIFY3D (19) and PROCHECK (20) do not have the capability to detect knots. We hope that with our contribution, the discovery of knots in newly solved protein structures becomes part of the standard routine, similar to identification of secondary structure elements or classification of protein's architecture.
To address this challenge, we developed a web server that allows a user to check a new or a known protein structure for knots by entering its PDB id or uploading a coordinate file.
MATERIALS AND METHODS
How knots are determined
Mathematically, knots are only well defined in closed (circular) loops (21). However, both the N- and C-termini of open proteins are typically located close to the surface of the protein and can be connected unambiguously: We reduce the protein to its backbone and draw two lines outward starting at the termini in the direction of the connection line between the center of mass of the backbone and the respective ends. The two lines are joined by a big loop, and the structure is topologically classified by the computation of its Alexander polynomial (21,22). To determine an estimate for the size of the knotted core, we successively delete amino acids from the N-terminus until the protein becomes unknotted (11). The procedure is repeated at the C-terminus starting with the last N-terminal deletion structure that contained the original knot. For each deletion, the outward-pointing line through the new termini is parallel to the respective lines computed for the full structure. Unfortunately, the size of a knot is not always precisely determined by this procedure, so reported sizes should only be regarded as approximate.
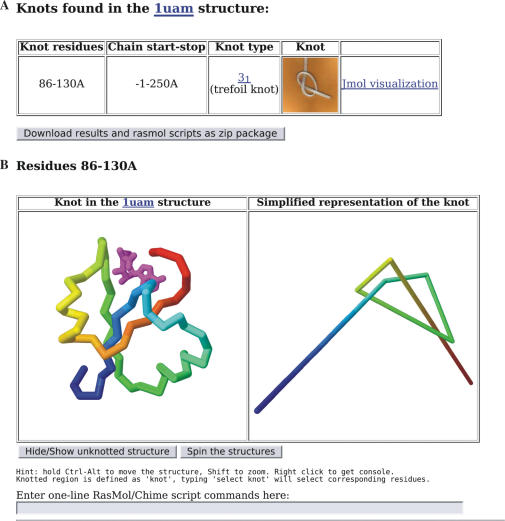
To speed up calculations, the KMT reduction scheme is used (9,11,23,24). This algorithm successively deletes amino acids that are not essential to the topological structure of the protein. It is also employed to create a reduced representation of the knot (Figure 1).
Figure 1.
The output of the Knots server for H. influenzae TrmD (PDB id 1uam). (A) Page one: the summary table. (B) Page two: Jmol interactive visualization. The 1uam structure is displayed in the left window with a knot highlighted in rainbow colors and the rest of the protein hidden. In this case, the trefoil knot spans a relatively small region of the protein and can be easily seen by eye in the protein structure. In many cases, this is difficult and the right panel offers the view of a simplified (reduced) representation of the knot. These visualizations can also be viewed offline using Rasmol scripts provided in the downloadable package.
In the course of our investigations (11) we came up with a number of stringent criteria that a structure should satisfy to be classified as knotted:
The Alexander polynomial should yield a knot.
There should not be any gaps in the polypeptide backbone. (See below.)
The knot should persist if two amino acids are removed from each end. (This prevents knots formed by just a few residues at the end of the chain passing through the loop—‘shallow knots’ and knots which only appear due to our specific loop-closure procedure.)
Unfortunately, there are some structures containing regions of the backbone that were not resolved and for which coordinates are not reported in PDB (a gap in the structure). Mobile loops may not be resolved by X-ray crystallography unless they are stabilized by a ligand or by protein engineering, for example. If the polypeptide chain contains a gap, the knot is reported if (i) a knot is present in at least one fragment of the chain and (ii) the structure that results from gaps being bridged with straight lines contains a knot. These criteria form the basis of our list of known knots. We have also included knotted structures with gaps if at least one homolog is knotted.
Server: input and output
As an input, the server accepts a structure file (in PDB or mmCIF format) or a PDB ID. The structure is tested for knots as described above. An option allowing a user to decide how to deal with the unresolved part of the structure is provided. The user may choose to connect unresolved parts by straight lines or to treat them as described above.
Figure 1 presents a typical output of the server—the summary page reporting a knot. If a knot is found, the server reports the type of the knot (e.g. 31- the trefoil knot, 41- the ‘figure eight’ knot, 52, etc.), its location in the protein structure, and a simplified representation of the knot (Figure1A). At this point, a user may choose to download results of the calculation as a collection of Rasmol/Jmol scripts or to proceed to the second page that has Jmol visualization of the knot on our server.
An option to download the tabulated results, the original structure file, and simplified structure and visualization Rasmol/Jmol scripts in one zip package is provided. This also can be used in cases when Jmol fails to start due to structure size or web browser-Java incompatibility issues.
The second page (Figure1B) has a two-window GUI to examine, rotate and further analyze the structure of the knot. The left window visualizes the protein structure with the knot using a Jmol Java applet. The knotted part is colored in rainbow colors to facilitate following the chain and visualizing the knot. The right window presents a simplified representation of the knot obtained by the reduction algorithm, making it easier to see that the protein structure is indeed knotted. The structures in both windows can be rotated, magnified and further analyzed using the tools of Jmol applet. Two buttons below (i) hide or show the rest of the protein structure in the left window, thus allowing a user to focus on the knot or to examine it in the context of the structure; and (ii) to spin structures in both windows simultaneously. An expert user familiar with Rasmol/Jmol commands can further analyze the structure using the command-line interface by entering individual commands or a whole script into a field below the windows.
The front page of the server also provides a curated list of discovered knots in proteins, classified according to the type of the knot, as well as a brief definition.
The server is implemented as a CGI Perl script, while the algorithmic part is written in C. The results of the calculation are stored for 20 min on the server, after which they have to be recomputed. Knot detection typically takes one to a few seconds.
Example of using the server
The bacterial tRNA(m1G37)methyltransferase (TrmD) is an enzyme that transfers methyl group from s-adenosyl-l-methionine (AdoMet) to a G nucleotide in the anti-codon region of certain bacterial tRNA species. The methylation of anti-codon nucleotides is essential for reducing the error rate in anti-codon binding to the complementary codon on mRNA during translation. The crystal structure of the enzyme from Haemophilus influenzae has recently been solved and is known to have a trefoil knot in the AdoMet-binding pocket (26). The specific configuration of the pocket allows AdoMet to adopt an unusual strongly bent conformation with its methyl group protruding from the pocket and accessible for transfer reaction (26).
The Knots output for a PDB entry 1uam, H. influenzae TrmD protein, is shown in Figure 1. A trefoil knot has been correctly identified for residues 86–130A in the 1uam structure. Clicking on ‘Jmol visualization’ link leads to the second page showing a protein ribbon diagram (Figure 1B, left), and the simplified representation of the knot. The knot can be easily seen in the protein structure by eye if the surrounding structure is hidden from view using the button provided.
The reduced representation of the knot (Figure 1B, right panel) is generated by the KMT reduction algorithm. The first and the last segments in this representation are not part of the protein but represent the connection lines to ‘infinity’, which are required to circularize the structure and calculate the Alexander polynomial (see the Section ‘How knots are determined’).
To ensure that the knot is not an artifact of connecting a gap in the structure, one may want to test for knots in each protein fragment separately. This option is provided on the front page of the server. In the case of the 1uam structure, the knot is found in one of the fragments.
More examples of knot in protein structure and their analysis can be found in our recent publication (11).
New protein knots discovered in 2006
Table 1 lists all novel protein knots that were discovered with our software in 2006. A complete list with all knotted proteins is available online.
Table 1.
Protein knots discovered in 2006
| Protein | Species | PDB code | Length | Type | Knotted core |
|---|---|---|---|---|---|
| α/β knot | Homo sapiens | 2ha8 | 159 | 31 | 103–148 (30) |
| Porphyromonas gingivalis | 2i6d | 231 | 31 | 177–222 (9) | |
| s-Adenosylmethionine synthetase | Homo sapiens | 2p02 | 380 | 31 | 59–302(21) |
| Ubiquitin hydrolase UCH-L1 | Homo sapiens | 2etl | 219 | 52 | 10–216 (7) |
Length refers to the size of the protein in amino acids. The knotted core is the minimum configuration that stays knotted after a series of deletions from each terminus; in parentheses we indicate how many amino acids can be removed from each side before the structure becomes unknotted. Note that unlike in our previous work (11), PDB residue numbers are used to describe the location of the knots.
CONCLUSION AND OUTLOOK
In this article, we presented our knot detection server and an illustration of its use. The server is easy to use, accurate and fast. In future, we plan to add automatic modeling of unresolved parts in the structures by using homology.
ACKNOWLEDGEMENTS
This work was supported by National Science Foundation grant DMR-04-26677 and by the Deutsche Forschungsgemeinschaft grant VI237/1. L.M. is an Alfred P. Sloan Research Fellow. Funding to pay the Open Access publication charges for this article was provided by the NIH-funded National Center for Biomedical Computing, Informatics for Integrating Biology and the Bedside (i2b2).
Conflict of interest statement. None declared.
REFERENCES
- 1.Liu LF, Depew RE, Wang JC. Knotted single-stranded DNA rings: a novel topological isomer of circular single-stranded DNA formed by treatment with Escherichia coli omega protein. J. Mol. Biol. 1976;106:439–452. doi: 10.1016/0022-2836(76)90095-4. [DOI] [PubMed] [Google Scholar]
- 2.Dean FB, Stasiak A, Koller T, Cozzarelli NR. Duplex DNA knots produced by Escherichia coli topoisomerase I. J. Biol. Chem. 1985;260:4975–4983. [PubMed] [Google Scholar]
- 3.Rybenkov VV, Cozzarelli NR, Vologodskii AV. The probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl Acad. Sci. USA. 1993;90:5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw SY, Wang JC. Knotting of a DNA chain during ring closure. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- 5.Arsuaga J, Vazquez M, Trigueros S, Sumners DW, Roca J. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc. Natl Acad. Sci. USA. 2002;99:5373–5377. doi: 10.1073/pnas.032095099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukin O, Vögtle F. Knotting and threading of molecules: chemistry and chirality of molecular knots and their assemblies. Angew. Chem. Int. Ed. 2005;44:1456–1477. doi: 10.1002/anie.200460312. [DOI] [PubMed] [Google Scholar]
- 7.Mansfield ML. Are there knots in proteins? Nat. Struct. Mol. Biol. 1994;1:213–214. doi: 10.1038/nsb0494-213. [DOI] [PubMed] [Google Scholar]
- 8.Mansfield ML. Fit to be tied. Nat. Struct. Mol. Biol. 1997;4:166–167. doi: 10.1038/nsb0397-166. [DOI] [PubMed] [Google Scholar]
- 9.Taylor WR. A deeply knotted protein structure and how it might fold. Nature. 2000;406:916–919. doi: 10.1038/35022623. [DOI] [PubMed] [Google Scholar]
- 10.Lua RC, Grosberg AY. Statistics of knots, geometry of conformations, and evolution of proteins. PLoS Comp. Biol. 2006;2:350–357. doi: 10.1371/journal.pcbi.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virnau P, Mirny LA, Kardar M. Intricate knots in proteins: function and evolution. PLoS Comp. Biol. 2006;2:1074–1079. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morizono H, Cabrera-Luque J, Shi D, Gallegos R, Yamaguchi S, Yu XL, Allewell NM, Malamy MH, Tuchman,M Acetylornithine transcarbamylase: a novel enzyme in arginine biosynthesis. J. Bacteriol. 2006;188:2974–2982. doi: 10.1128/JB.188.8.2974-2982.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi D, Morizono H, Aoyagi M, Tuchman M, Allewell NM. Crystal structure of human ornithine transcarbamylase complexed with carbamyl phosphate and L-norvaline at 1.9 A resolution. Proteins: Struct. Funct. Genet. 2000;39:271–277. [PubMed] [Google Scholar]
- 14.Das C, Hoang QQ, Kreinbring CA, Luchansky SJ, Meray RK, Ray SS, Lansbury PT, Ringe D, Petsko GA. Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1. Proc. Natl Acad. Sci. USA. 2006;103:4675–4680. doi: 10.1073/pnas.0510403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misaghi S, Galardy PJ, Meester WJN, Ovaa H, Ploegh HL, Gaudet,R Structure of the ubiquitin hydrolase Uch-L3 complexed with a suicide substrate. J. Biol. Chem. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 16.Mallam AL, Jackson SE. Folding studies on a knotted protein. J. Mol. Biol. 2005;346:1409–1421. doi: 10.1016/j.jmb.2004.12.055. [DOI] [PubMed] [Google Scholar]
- 17.Mallam AL, Jackson SE. Probing nature's knots: the folding pathway of a knotted homodimeric protein. J. Mol. Biol. 2006;359:1420–1436. doi: 10.1016/j.jmb.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 18.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 19.Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 21.Adams CC. The Knot Book: An Elementary Introduction to the Mathematical Theory of Knots. New York: Freeman; 1994. [Google Scholar]
- 22.Virnau P, Kantor Y, Kardar M. Knots in globule and coil phases of a model polyethylene. J. Am. Chem. Soc. 2005;127:15102–15106. doi: 10.1021/ja052438a. [DOI] [PubMed] [Google Scholar]
- 23.Virnau P, Kardar M, Kantor Y. Capturing knots in polymers. Chaos. 2005;15:041103. doi: 10.1063/1.2130690. [DOI] [PubMed] [Google Scholar]
- 24.Koniaris K, Muthukumar M. Self-entanglement in ring polymers. J. Chem. Phys. 1991;95:2873–2881. [Google Scholar]