Abstract
The analysis of expressed sequence tag (EST) datasets offers a rapid and cost-effective approach to elucidate the transcriptome of an organism, but requiring several computational methods for assembly and annotation. ESTExplorer is a comprehensive workflow system for EST data management and analysis. The pipeline uses a ‘distributed control approach’ in which the most appropriate bioinformatics tools are implemented over different dedicated processors. Species-specific repeat masking and conceptual translation are in-built. ESTExplorer accepts a set of ESTs in FASTA format which can be analysed using programs selected by the user. After pre-processing and assembly, the dataset is annotated at the nucleotide and protein levels, following conceptual translation. Users may optionally provide ESTExplorer with assembled contigs for annotation purposes. Functionally annotated contigs/ESTs can be analysed individually. The overall outputs are gene ontologies, protein functional identifications in terms of mapping to protein domains and metabolic pathways. ESTExplorer has been applied successfully to annotate large EST datasets from parasitic nematodes and to identify novel genes as potential targets for parasite intervention. ESTExplorer runs on a Linux cluster and is freely available for the academic community at http://estexplorer.biolinfo.org.
INTRODUCTION
Expressed sequence tags (EST) represent short, unedited, randomly selected single-pass sequence reads derived from cDNA libraries, providing a low-cost alternative (also called ‘poor’ man's genome) to whole genome sequencing (1,2) and specifically relevant to the transcriptome of an organism at various stages of development or under different experimental conditions. The analysis of EST data can enable gene discovery, complement genome annotation, aid gene structure identification, establish the viability of alternative transcripts, guide single nucleotide polymorphism (SNP) characterization and facilitate proteomic exploration (2). ESTs are highly error prone and require several computational methods for pre-processing, clustering, assembly and annotation to yield biological information. Furthermore, it is extremely important to be able to store, organize and annotate ESTs using a comprehensive analysis pipeline due to their ‘high-throughput’ nature.
We recently compared (2) available web resources (http://biolinfo.org/EST/), individual tools and pipelines pertaining to EST analysis. We also evaluated currently available methods for each step of analysis, including EST clustering, assembly, consensus generation and tools for DNA and protein annotation, employing benchmark EST datasets. A detailed investigation of different EST analysis platforms (3–8) revealed that they all terminate prior to functional annotations, such as gene ontologies, motif/pattern analysis and pathway mapping. Some platforms terminate at the assembly level, providing contigs and singletons as an output (3). Other platforms solely run nucleotide-based programs with limited annotation at the protein level (5,7,9,10). Therefore, we developed ESTExplorer, a complete EST analysis suite which employs programs for both nucleotide- and protein-based annotation. Moreover, we have carefully selected the most appropriate combination of programs for each stage of EST analysis, based on their ability to accurately reproduce partial gene sequences from ESTs and annotate them as correctly as possible (http://estexplorer.biolinfo.org/methodology.html).
ESTExplorer comprises a suite of programs with a customizable web interface to manage and analyse EST data. Optionally, EST assembly datasets generated elsewhere, e.g. EGAssembler (3), can be further functionally annotated at the ESTExplorer website. Users have the option of selecting specific analysis phases (detailed below). Besides pre-processing and assembly from EST sequences, ESTExplorer annotates input sequences extensively, using gene ontologies (GO), domain analysis and pathway mapping. ESTExplorer has been used extensively for the analysis and annotation of large EST datasets from parasitic nematodes generated in our laboratories, and to identify key nematode molecules as potential targets for anti-parasite intervention. ESTExplorer has been also used for the analysis of differential transcription between adult male and female Haemonchus contortus by oligonucleotide microarrays (unpublished data).
OVERVIEW OF ESTEXPLORER
The ESTExplorer workflow can be divided into three phases (shown in Supplementary Figure 1). Phase I is dedicated to EST sequence pre-processing and assembly, Phase II carries out DNA- level annotation and Phase III provides for protein-level annotation.
ESTExplorer can accept nucleotide sequence input of two types (Figure 1A; arrows in Supplementary Figure 1). ESTs in FASTA format can be submitted to Phase I for EST pre-processing and assembly, followed by analyses in Phases II and III. Alternatively, ESTs assembled using another program or pipeline into contigs and singletons, may be submitted directly for functional annotation (Phases II and III).
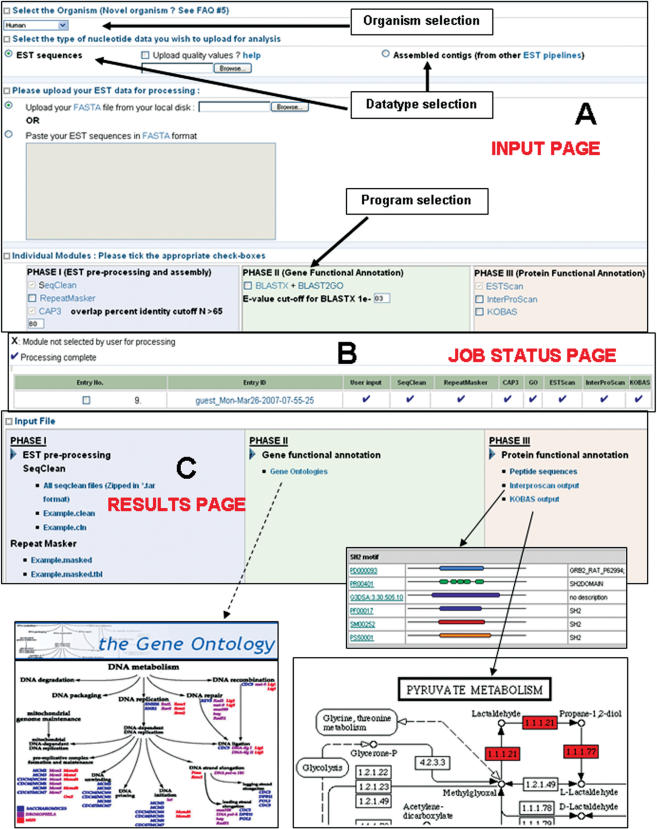
Figure 1.
ESTExplorer input and analysis output pages. (A) EST or contig submission with optional parameters for organism and program selection, (B) Status page to monitor the progress of different programs and (C) Result download page from where processed data can be downloaded and annotation links accessed. Screenshots of the results showing Gene Ontologies (accessed via a link for each EST/contig), InterProScan and KEGG pathway mapping obtained from processed EST data are shown.
Phase I comprises three programs run sequentially, to convert input EST sequences into high quality ESTs. SeqClean accepts ESTs in FASTA format and performs vector removal (using NCBI's UniVec database), PolyA removal, trimming of low quality segments at the 5′ and 3′ ends and cleaning of low complexity regions (using the DUST module). Additionally, all short ESTs (<100 bp) are eliminated as uninformative. The output from SeqClean is processed by RepeatMasker (11) to mask repeats. Species-specific repeat masking is done using Repeat Masker which in turn employs Cross_Match and up-to-date repeat libraries for different species from RepBase. For a novel species, the nearest organism listed in ESTExplorer, using NCBI Taxonomy, may be selected. CAP3 (12) then accepts repeat-masked high quality EST sequences and performs clustering and assembly into contigs (containing multiple ESTs) and singletons, based on an overlap percent identity threshold cutoff of 80. The user can modify this, with the recommendation to provide a value >65. Output files from each program are provided.
Phase II carries out annotation at the nucleotide level, of assembled EST contigs and singletons from Phase I or directly uploaded by the user, using the BLASTX (13) program and NCBI's non-redundant protein database, followed by the assignment of functionality via Gene Ontologies (14) using BLAST2GO (15). BLAST2GO extracts GO terms for each BLAST hit obtained by mapping to extant annotation associations, using a default cutoff of E-03, which the user can modify. Additionally, BLAST2GO provides a data file which can be used to reconstruct GO relationships and perform statistical analysis on gene function information. ESTExplorer, in turn, retrieves gene ontologies from BLAST2GO and links each GO identifier to its ontology tree, displayed by the AmiGO Browser.
Protein-based annotation is effected in Phase III. At the outset, ESTScan (16) accepts contigs and singletons from CAP3 and provides conceptual translations, using the genetic code from the nearest organism, in a two-step process. In the first step, coding regions or open reading frames (ORFs) are detected and extracted, while correcting for frame shift errors. In the second step, these ORFs are translated into putative peptides. ESTExplorer currently implements the genetic codes (smat files generated from mRNA sequences) for the ten organisms: human, mouse, rat, rice, zebrafish, chicken, fly, dog, thale cress (Arabidopsis thaliana) and roundworm (Caenorhabditis elegans) provided by the authors of ESTScan. For a novel species, the nearest organism listed in ESTExplorer, using NCBI Taxonomy, may be selected. The peptide sequences from ESTScan are simultaneously passed on to InterProScan (17) and KOBAS (18) for processing. InterProScan matches protein sequences against InterPro, an integrated resource for protein families, domains and functional sites from member databases such as PROSITE, PRINTS, Pfam, ProDom and SMART. ESTExplorer runs InterProScan in the backend and provides an html output that users can download and analyse, with details of domain/motif architecture for each sequence. KOBAS (KEGG orthology-based annotation system) maps protein sequences to pathways based on KEGG (19). KOBAS uses controlled vocabularies (KO) to annotate a set of sequences and assigns pathways to individual proteins, using a two-step process. In the first step, it takes a set of sequences and assigns KEGG orthology terms based on a BLASTP similarity search against KEGG GENES or direct cross-sequence identifier mapping. In the second step, KO is used for respective pathway identification. ESTExplorer provides an html output for the mapped pathways through which the user can directly access the pathways at the KEGG website. Proteins that are mapped from the processed EST dataset are highlighted and coloured differently for easy identification.
Once an EST or contig dataset has been submitted to ESTExplorer, a status page is accessible (Figure 1B), for monitoring the progress of the analysis, at the program level. As each selected program is completed, the status page is updated and the output from that program becomes available immediately.
ESTExplorer provides an integrated workflow approach to EST analysis, by combining assembly with traditional and well-established resources, such as BLAST2GO and InterPro. While some components are available separately as web servers, ESTExplorer has extended functionality over these as well as added additional features, interfaced seamlessly together. Phase I of ESTExplorer roughly maps to the functionality of EGAssembler. However, there is no functional annotation after assembly into contigs from EGAssembler. Additionally, we have also provided the ability to use quality values during the assembly process. Phase II involves DNA-level orthologue mapping, directly from the Phase I output. When there are several contigs and singletons after Phase I, the user does not have to submit each one to NCBI to run BLAST. Additionally, we can process each of the contigs and singletons from the Phase I for protein-level annotation, via Phase III where the complete InterproScan, GO mapping and KEGG pathway mapping are carried out. Recently, Pavy and coworkers (20) have used GO and Pfam matches for annotating their ESTs at a functional level. ESTExplorer provides these along with the additional advantage of KEGG and the complete InterProScan currently comprising 12 modules in addition to Pfam, for protein and domain analysis (details available from our website).
The outcome for each run is summarized, with links to output files from each selected program. An email with the URL of the results will be sent to the user after the completion of the entire run. Users can either download output files from the download page for each step or as a single zipped file for each phase of the analysis (Figure 1C). The results are stored for one week, after the completion of the run. Some programs are run by default, whereas others are optional. In Phase I, SeqClean and CAP3 are run by default while RepeatMasker is optional. All of the programs in Phase II and III, excepting ESTScan, are optional. We update the backend databases (non-redundant protein and UniVec databases from NCBI, Repeat Database from RepBase, Gene Ontologies, InterProScan and KEGG) every month using automated scripts. A detailed tutorial and FAQ (http://estexplorer.biolinfo.org/tutorial.html) are available for running sample EST datasets and understanding the different analysis programs.
It is usually difficult to collate the analysis results at the final output stage when a large dataset is analysed using a workflow containing several phases and multiple programs. To address this issue, ESTExplorer tracks each assembled sequence (contig/singleton) which has been functionally annotated (more details are available from the example section).
SOFTWARE/HARDWARE ENVIRONMENT
ESTExplorer has been developed using open source technologies; Zope (V2.8.1), Python (V2.4.3) and MySQL (V4.1.10a), for EST data management and analysis. ESTExplorer runs on a 16-node Linux cluster (1.3 GHz, Itanium 2 Rev, 5 Processors, 16 GB RAM) running on Red Hat Enterprise Linux AS Release 3. The workflow architecture has been designed based on a ‘distributed control approach’. The user request from the central ZOPE controller is diverted to one of the data-processing machines after appropriate load balancing. Browser and platform independent java scripts have been used for data validation, in order to enhance the flexibility of query and output pages. The server refreshes the intermediate result page every 30s and updates the user with the status of processing in the individual programs in the pipeline. A final output page provides the user with detailed output files for viewing and for downloading the results. Output files are stored on the server for seven days.
EXAMPLES OF APPLICATIONS
From dbEST (21), we provide a small dataset of 372 ESTs (Input Option 1 in Supplementary Figure 1) for the plant Capsicum chinense and the complete analysis results from ESTExplorer. Additionally, assembled sequences (contigs/singletons) from these ESTs have been provided as an example for Input Option 2 (Supplementary Figure 1). Detailed sequence-wise annotation summaries are provided to facilitate rapid functional analysis of EST datasets (http://estexplorer.biolinfo.org/example_capsicum/summary_table.html). The detailed summary of the analysis of contig 9 shows the contributing ESTs, protein domains, gene ontologies and mapped pathway (shown in Supplementary Figure 2).
One of our research projects involves gene discovery from parasitic nematodes. ESTExplorer has allowed the rapid and accurate analysis of ESTs by providing robust annotation at the gene and protein levels, matching evidence from multiple sources. Using ESTExplorer to analyse 873 ESTs from a parasitic nematode Oesophagostomum dentatum (22) yielded 133 contigs and 314 singletons, compared with 128 contigs and 388 singletons reported by Cottee et al. (22). Overall, 29 entries were annotated with gene ontology data, 44 sequences had protein domain information and 246 sequences were mapped to KEGG pathways. This rapid and comprehensive analysis together with additional analyses of specific molecules enabled the identification of novel genes and molecules predicted, based on comparisons with extensive data in WormBase (23), to be involved in biological pathways critical for development, reproduction and survival. With ESTExplorer, the analysis was systematic and additional information on domain and pathway mapping made it easier to validate functional annotation with low scoring hits. This dataset is provided as the second example dataset (Input Data 1) on the server (http://estexplorer.biolinfo.org/examples.html). A moderate dataset of 10 651 ESTs for Ancylostoma ceylanicum, downloaded from dbEST (21), is also available, as ESTs in FASTA format (Input Option 1) and assembled ESTs (Input Option 2).
Additionally, we have also applied ESTExplorer for the analysis of a number of EST datasets ranging from 717 ESTs from a related parasitic nematode Trichostrongylus vitrinus (24) to 21 967 ESTs from Haemonchus contortus for subsequent analysis of differential transcription between adult male and female worms by oligonucleotide microarrays. We used two types of data that were annotated using ESTExplorer: the first comprised unprocessed 21 967 ESTs and the second contained 1885 contigs. By annotating both the ESTs as well as these contigs, we have been able to get better representation of biologically relevant genes for oligonucleotide design and subsequent microarray analysis (unpublished data). ESTExplorer has been used extensively for the annotation of transcript and protein sequence data for the Aspergillus niger and Mycosphaerella graminicola fungal genomes, a collaborative effort of our group (N.D. and S.R.) with DOE Joint Genome Institute (JGI), USA.
FUTURE DIRECTIONS
ESTExplorer currently supports organism-based repeat masking and conceptual translation for ten commonly researched model organisms per se. Our goal is to extend this capability to several newly sequenced organisms. In this direction, we are adding data for additional species for repeat masking and conceptual translation. Users will also be able to upload their own data files during pre-processing (vectors, adaptors, organism-specific repeats) and their own databases for similarity searches, for the targeted analysis of EST sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Michael Baxter, Macquarie University, Australia; Gary Cobon, Genetic Technologies Limited, Australia; Ana Conesa and Stefan Goetz, Centro de Genómica, Spain; Christian Iseli, SIB, Switzerland; Sarah Hunter, EBI, UK and Xizeng Mao, Peking University, China for their invaluable help and support. We are grateful to Macquarie University for the award of iMURS research scholarships (S.H.N. and N.D.) and an MUPGR travel grant (S.H.N.) and to the Macquarie University Biotechnology Research Institute for the award of a Ph.D. top-up scholarship (S.H.N.). Partial support from Genetic Technologies Limited and the Australian Research Council (LP0667795) are acknowledged. Funding to pay the Open Access publication charges for this article was provided by Macquarie University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, Utterback TR, Nagle JW, Fields C, Venter JC. Sequence identification of 2,375 human brain genes. Nature. 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 2.Nagaraj SH, Gasser RB, Ranganathan S. A hitchhiker's guide to expressed sequence tag (EST) analysis. Brief Bioinform. 2007;8:6–21. doi: 10.1093/bib/bbl015. [DOI] [PubMed] [Google Scholar]
- 3.Masoudi-Nejad A, Tonomura K, Kawashima S, Moriya Y, Suzuki M, Itoh M, Kanehisa M, Endo T, Goto S. EGassembler: online bioinformatics service for large-scale processing, clustering and assembling ESTs and genomic DNA fragments. Nucleic Acids Res. 2006;34:W459–W462. doi: 10.1093/nar/gkl066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao C, Cushman JC, May GD, Weller JW. ESTAP – an automated system for the analysis of EST data. Bioinformatics. 2003;19:1720–1722. doi: 10.1093/bioinformatics/btg205. [DOI] [PubMed] [Google Scholar]
- 5.Hotz-Wagenblatt A, Hankeln T, Ernst P, Glatting KH, Schmidt ER, Suhai S. ESTAnnotator: a tool for high throughput EST annotation. Nucleic Acids Res. 2003;31:3716–3719. doi: 10.1093/nar/gkg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkinson J, Anthony A, Wasmuth J, Schmid R, Hedley A, Blaxter M. PartiGene – constructing partial genomes. Bioinformatics. 2004;20:1398–1404. doi: 10.1093/bioinformatics/bth101. [DOI] [PubMed] [Google Scholar]
- 7.D'Agostino N, Aversano M, Chiusano ML. ParPEST: a pipeline for EST data analysis based on parallel computing. BMC Bioinformatics. 2005;6(Suppl. 4):S9. doi: 10.1186/1471-2105-6-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, Lee Y, White J, Cheung F, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 9.Latorre M, Silva H, Saba J, Guziolowski C, Vizoso P, Martinez V, Maldonado J, Morales A, Caroca R, et al. JUICE: a data management system that facilitates the analysis of large volumes of information in an EST project workflow. BMC Bioinformatics. 2006;7:513. doi: 10.1186/1471-2105-7-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paquola AC, Nishyiama MY, Jr, Reis EM, da Silva AM, Verjovski-Almeida S. ESTWeb: bioinformatics services for EST sequencing projects. Bioinformatics. 2003;19:1587–1588. doi: 10.1093/bioinformatics/btg196. [DOI] [PubMed] [Google Scholar]
- 11.Smit A. 2007. [accessed on 20/01/2007.]. Repeat Masker http://www.repeatmasker.org/
- 12.Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 16.Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1999;7:138–148. [PubMed] [Google Scholar]
- 17.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavy N, Paule C, Parsons L, Crow JA, Morency MJ, Cooke J, Johnson JE, Noumen E, Guillet-Claude C, et al. Generation, annotation, analysis and database integration of 16,500 white spruce EST clusters. BMC Genomics. 2005;6:144. doi: 10.1186/1471-2164-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boguski MS, Lowe TM, Tolstoshev CM. dbEST – database for “expressed sequence tags”. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 22.Cottee PA, Nisbet AJ, Abs El-Osta YG, Webster TL, Gasser RB. Construction of gender-enriched cDNA archives for adult Oesophagostomum dentatum by suppressive-subtractive hybridization and a microarray analysis of expressed sequence tags. Parasitology. 2006;132:691–708. doi: 10.1017/S0031182005009728. [DOI] [PubMed] [Google Scholar]
- 23.Bieri T, Blasiar D, Ozersky P, Antoshechkin I, Bastiani C, Canaran P, Chan J, Chen N, Chen WJ, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nisbet AJ, Gasser RB. Profiling of gender-specific gene expression for Trichostrongylus vitrinus (Nematoda: Strongylida) by microarray analysis of expressed sequence tag libraries constructed by suppressive-subtractive hybridisation. Int. J. Parasitol. 2004;34:633–643. doi: 10.1016/j.ijpara.2003.12.007. [DOI] [PubMed] [Google Scholar]