Abstract
We sequenced the genome of Saccharomyces cerevisiae strain YJM789, which was derived from a yeast isolated from the lung of an AIDS patient with pneumonia. The strain is used for studies of fungal infections and quantitative genetics because of its extensive phenotypic differences to the laboratory reference strain, including growth at high temperature and deadly virulence in mouse models. Here we show that the ≈12-Mb genome of YJM789 contains ≈60,000 SNPs and ≈6,000 indels with respect to the reference S288c genome, leading to protein polymorphisms with a few known cases of phenotypic changes. Several ORFs are found to be unique to YJM789, some of which might have been acquired through horizontal transfer. Localized regions of high polymorphism density are scattered over the genome, in some cases spanning multiple ORFs and in others concentrated within single genes. The sequence of YJM789 contains clues to pathogenicity and spurs the development of more powerful approaches to dissecting the genetic basis of complex hereditary traits.
Keywords: comparative genomics, genome architecture, introgression, lateral gene transfer
There is extensive genetic and phenotypic diversity within species. Determining which of the vast amounts of sequence differences that are found among individuals of a species contribute to heritable traits will allow diseases to be tackled at the molecular level and aid in the development of novel therapies. Saccharomyces cerevisiae, commonly known as baker's or brewer's yeast, plays a central role in food production and is one of the most studied genetic model species. It is not only widely used in biotechnology but also is a powerful model system that has been applied to identify multigenetic factors of hereditary traits (1–7). The genome sequence of one laboratory strain, a derivative of S288c, was the first genome of a free-living eukaryotic organism to be sequenced (8). Over the last 10 years, this genome has served as the reference for the S. cerevisiae species and has catalyzed the development of whole-genome approaches to biology (9, 10). Despite frequent laboratory use of alternative strains, sequence information for S. cerevisiae beyond the domesticated strain S288c has been fragmentary.
S288c, which originated from a strain isolated from a rotten fig, was chosen for sequencing because it possesses properties that make it easy to work with, such as minimal colony morphology switching, consistent growth rates in glucose media, and no flocculence (11). At several loci, S288c contains polymorphisms not found in natural isolates, which could be hallmarks of domestication (12, 13). A growing number of S. cerevisiae infections in humans have recently been reported (14). As a result, S. cerevisiae is also regarded as an emerging opportunistic pathogen that can cause clinically relevant infections in different patient types and body sites (15–17). One clinical strain (YJM145), derived from a yeast isolated from an AIDS patient with S. cerevisiae pneumonia (18), has been studied extensively as a model for fungal infections (19–24). YJM145 causes death in complement-deficient mice (20), and its haploid isoform, YJM789, differs in phenotype from S288c, for example, in being flocculant, displaying colony morphology switching, and growing at high temperature. The high-temperature growth phenotype of YJM789 in particular has been dissected to an SNP resolution for several local regions of divergence (1, 7). Globally high divergence at the sequence level has been inferred from genetic crosses (19), from sequencing portions of its genome (1, 25), and from hybridization to oligonucleotide arrays that could detect the presence of SNPs and insertions/deletions (indels) but not their sequence identity (1, 25–28). Here we analyze the genome of YJM789 and compare it to S288c. The sequence of YJM789 has implications for the functional significance of genetic variation during pathogenicity, its evolutionary history, and the development of new approaches that determine the contribution of allelic variants to phenotypes.
Results and Discussion
Genome and Comparison.
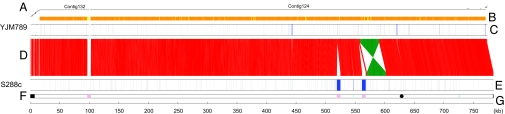
We sequenced the genome of strain YJM789 by using a shotgun approach, generating >170,000 sequence reads, followed by finishing to close gaps of the nonrepetitive portions of the genome, which yielded an additional ≈4,000 reads. After assembly, 11.8 Mb of high-quality genome sequence were obtained. The coverage corresponds to 98% of the S288c genome as determined from chromosome-by-chromosome alignments of the two genome sequences [see Fig. 1 for chromosome XIV and supporting information (SI) Fig. 5 for the entire genome]. The 16 YJM789 nuclear chromosomes are covered by 31 contigs (see SI Tables 1 and 2) and the mitochondrial genome (mtDNA) by a single contig.
Fig. 1.
Alignment of the chromosome XIV sequences from YJM789 and S288c. (A) YJM789 contigs mapped to their locations on the S288c genome. (B) Sequence similarity between YJM789 contigs with a length of at least 10 kb and their corresponding sequences of S288c represented by color and coded from yellow (low) to orange (high). (C) Sequences of ≥100 bp that are present in YJM789 but absent in S288c are represented by blue lines. Similar sequences of <100 bp are represented by gray lines. (D) Sequence alignment between YJM789 and S288c chromosome XIV. Identical sequences are linked by lines. Red represents forward alignment. Green represents reverse complementary alignment. (E) Sequences of ≥100 bp that are absent in YJM789 but present in S288c are represented by blue lines. Similar sequences of <100 bp are represented by gray lines. (F) Repeat sequences of S288c are represented as follows: cyan rectangles, long terminal repeats; pink rectangles, retrotransposons; black rectangles, telomeres; black circle, centromere. (G) Coordinates of S288c in kilobase pairs.
ORFs and Horizontal Transfer.
Employing three methods, we predicted 5,904 ORFs in the nuclear genome of YJM789, of which 5,509 have a reciprocal-best-hit ortholog in S288c (see SI Data Set 1 for a list of all ORFs, plus notes and descriptions). Several of the potentially unique YJM789 ORFs have a nonreciprocal-best-hit homolog in S288c (114), whereas several others have near-perfect-match hits in nucleotide sequence to S288c (116) and likely reflect ORF annotation differences. However, 165 ORFs are predicted to be absent in S288c because of early stop codons or the absence of start codons, generated by SNPs or indels. In addition, the YJM789 genome has some ORFs whose sequences are not at all present in the laboratory strain (see SI Data Set 1 for a complete list of these ORFs and comments). One such example is yorf4.16.070.031, encoding a hypothetical protein that is part of a 3.77-kb region unique to YJM789 (SI Fig. 6). Another unique YJM789 ORF is recognizable as KHR1, encoding a heat-resistant killer toxin (29). KHR1 is located in a 1.94-kb sequence unique to YJM789 that is flanked by direct repeats of a Ty element. At the corresponding position in S288c, there is only one Ty element (YILCdelta3), suggesting that a recombination between the direct repeats may be responsible for the absence of KHR1 in some S. cerevisiae strains, including S288c.
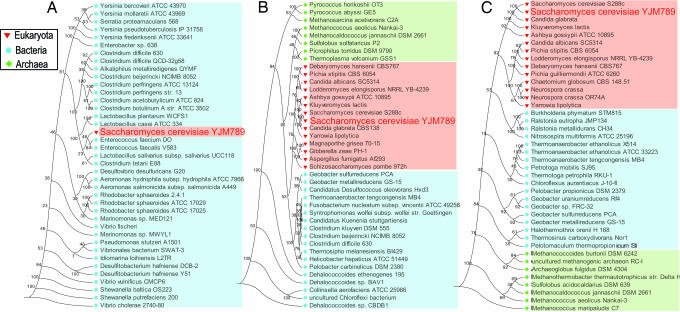
Two further examples of genes that are present in YJM789 but absent in S288c are RTM1, which encodes a protein that confers resistance to molasses (30), and one unknown gene. The presence of these two genes in YJM789 was confirmed by PCR amplification and sequencing. With regard to the unknown gene, BLASTP against GenBank was performed by using its amino acid sequence. The BLAST results contain mostly GCN5-related N-acetyltransferases (GNAT) from different bacteria, with the top hit being an uncharacterized protein from Enterococcus faecium strain DO; therefore, we named this unknown gene YJM-GNAT. Some members of the GNAT superfamily are known to confer resistance to aminoglycoside antibiotics in certain bacteria, such as E. faecium (31, 32) and Salmonella enterica (33). Furthermore, the phylogeny for YJM-GNAT differs dramatically from phylogenies obtained for other YJM789 genes (see Fig. 2). Although further analysis will be informative to completely rule out the possibility of gene loss (34), these data suggest that YJM-GNAT might have been transferred horizontally from a bacterium.
Fig. 2.
Phylogenetic tree of YJM-GNAT homologs. (A) YJM-GNAT homologs were retrieved by BLASTP against the nonredundant database by using the threshold (E value of ≤1 × 10−5, identity of ≥30%, and the alignment matching at least 75% of the length of both query and subject sequences). Representative species are shown. Multiple alignments were built by CLUSTALW. A phylogenetic tree was then constructed by using the neighbor-joining method of the PHYLIP package. GNAT homologs are represented by their species names. (B and C) Phylogenies of two other YJM789 genes encoding acetyltrasferases for comparison: ELP3 (B) and ECM40 (C).
Inversion and Translocation.
A chromosome-by-chromosome sequence comparison of the YJM789 and S288c genomes shows one large inversion (32.5 kb). The inversion spans the interval between base pairs 569,858 and 602,396 on chromosome XIV in S288c (base pairs 456,203–488,724 in contig 124 of YJM789) and is flanked by inverted repeats of ≈4.2 kb (Fig. 1). The presence of the inverted repeat sequences suggests a mechanism for the inversion: The repeats recombined with each other. Independent PCR analysis of genomic DNA from YJM789 and S288c corroborated the inversion. Analogous analysis of the vineyard isolate RM11-1a (35) and a sequence comparison to Saccharomyces paradoxus, the closest species to S. cerevisiae that has its genome sequenced (36), shows that, in both YJM789 and RM11-1a, this region is inverted relative to S288c and S. paradoxus. In addition, a translocation was detected between chromosomes VI and X, wherein an element of 18 kb on chromosome VI in S288c (base pairs 11,626–30,088) is found on chromosome X in YJM789 (base pairs 1–18,478 on contig 100). This translocation was confirmed independently by PCR amplification across the breakpoints and by sequencing the ends of the amplicons.
Highly Polymorphic Chromosomal Regions.
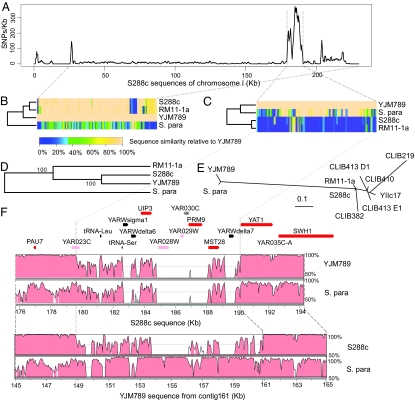
Within the aligned regions of the YJM789 and S288c genomes, we identified 59,361 high-confidence SNPs that are scattered throughout the genome (SI Table 3 and SI Fig. 7 present the SNP distribution for each individual chromosome). SNP density is 6.1 per kilobase on average but is far from constant across the genome and across individual chromosomes, with chromosome I having the highest average SNP density of 19.7 per kilobase. There is a discrete region of ≈12 kb on chromosome I that is highly polymorphic (Fig. 3). The abrupt transitions from low-to-high and high-to-low SNP density at its boundaries prompted us to analyze this chromosome I region in more detail.
Fig. 3.
Highly polymorphic region on chromosome I. (A) SNP distribution between YJM789 and S288c determined from a 1-kb sliding window over the nonrepeat sequence of S288c chromosome I. (B) Clustergram of the sequence similarity of chromosomes I of YJM789 compared with S288c, RM11-1a, and S. paradoxus (S. para) using a 1-kb sliding window. (C) Clustergram of the sequence similarity of the high polymorphism region of YJM789 chromosome I compared with S288c, RM11-1a, and S. paradoxus using a 100-bp sliding window. (D) Phylogeny of chromosome I sequences excluding the interval containing the high polymorphism region. The phylogenetic tree was constructed from nucleotide sequence alignments generated by using the program VISTA and the neighbor-joining method of the PHYLIP package. (E) Phylogeny of DUP240 region from YARWdelta6 to YARWdelta7 in S. paradoxus and all sequenced S. cerevisiae strains (38). A phylogenetic tree was constructed from nucleotide sequence alignments generated by CLUSTALW and the neighbor-joining method of the PHYLIP package. The scale bar indicates the evolutionary distance (number of substitutions per nucleotide position). (F) Alignments of YJM789, S288c, and S. paradoxus over the high polymorphism region using S288c (Upper) or YJM789 (Lower) as the reference sequence. The y axis represents the sequence similarity between two genomes along the reference sequence (graphs generated in VISTA). Sequence identity is shown for each pairwise comparison in a 100-bp sliding window. Note that differences in sequence lengths arise because of indels between YJM789 and S288c. Genes, as encoded in S288c, are represented by colored boxes: red, verified ORFs; pink, uncharacterized ORFs; gray, dubious ORFs; black, tRNAs and long terminal repeats.
Close examination of the highly polymorphic region on chromosome I (Fig. 3A) showed that the ≈12-kb sequence contains 2,356 SNPs and 187 indels, accounting for >50% of the total chromosome I polymorphisms. The region in S288c encompasses five members of the nonessential DUP240 gene family encoding putative integral membrane proteins (37): UIP3, YAR028W, YAR029W, PRM9, and MST28. The corresponding region of YJM789 contains the orthologs for UIP3, YAR028W, PRM9, MST28, as well as an ORF (yorf4.01.161.113) unique to YJM789. Recurrent deletion and ectopic recombination has been suggested to underlie the diversity of the DUP240 gene family regions among S. cerevisiae strains (38).
We compared the sequence of this region from S288c, RM11-1a, YJM789, the sibling species S. paradoxus, and six of the S. cerevisiae strains previously examined (Fig. 3 B–F) (38). Phylogenetic analysis of the DUP240 region on chromosome I shows that YJM789 is markedly distinct from all other S. cerevisiae strains but is similar to S. paradoxus (Fig. 3 C and E). As determined from a sequence outside this region on chromosome I, a different phylogenetic relationship exists among the strains (Fig. 3D). Indeed, the nucleotide similarity between YJM789 and S. paradoxus within the region appears higher (93%) than along the rest of chromosome 1 (Fig. 3B). Although several large indels exist between YJM789 and S. paradoxus, S. paradoxus is the organism with the highest similarity to YJM789 currently in the GenBank database. The average sequence identity is 85% between these two genomes (Fig. 3F).
Although rearrangements characterize variation in DUP240 ORFs among several S. cerevisiae strains (38), the degree of separation of YJM789 from other S. cerevisiae strains, the similarity between YJM789 and S. paradoxus, and the difference in phylogeny to genes outside this region were unexpected. Introgression between YJM789 and S. paradoxus or a closely related species is a possibility that can account for these observations. Indeed, S. paradoxus and S. cerevisiae share similar habitats (39), and hybrids between the two are found in nature (40). Although hybrids between S. cerevisiae and other members of the Saccharomyces sensu stricto are predominantly sterile, rare viable offspring containing DNA from both species have been produced (41–43), providing a putative way for introgression to occur.
Highly Polymorphic ORFs.
Regions of high polymorphism density also are found localized to individual ORFs. The absence of YJM789 DNA hybridization to oligonucleotides representing several S288c ORFs has been reported and, in some cases, interpreted as missing sequences rather than highly polymorphic regions (26). The genome sequence enables an investigation of this issue. We have obtained high-quality sequences covering the vicinities of the proposed ORF regions (SI Table 4). Six of these ORFs indeed appear to be absent in YJM789 (YHR054C, YIL080W, YIL082W, YIL082W-A, YJL113W, and YJL114W). However, 22 ORFs are confirmed to be present but highly polymorphic in YJM789, including six genes in the highly polymorphic chromosome I region.
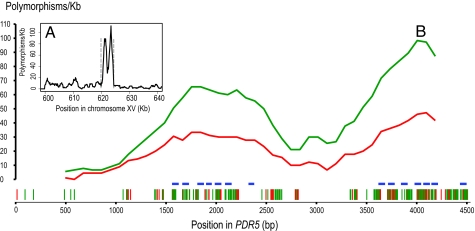
One notable example of a highly polymorphic ORF is PDR5, which encodes a multidrug transporter. PDR5 is among the genes predicted absent from the YJM789 genome (26). Sequencing shows that it is present but that it contains >250 SNPs (no indels), resulting in 5.3% amino acid differences between YJM789 and S288c. Because the regions flanking the PDR5 ORF are similar between both strains, the diverged region is highly localized (Fig. 4).
Fig. 4.
Polymorphism density across PDR5 between YJM789 and S288c. (A) Polymorphism distribution on chromosome XV from kilobases 600 to 640. Dashed lines indicate the start and stop positions of the PDR5 ORF. (B) The distribution of nonsynonymous and synonymous substitutions within the PDR5 ORF as determined from a 900-bp sliding window (each slide is 90 bp). The possibilities for nonsynonymous and synonymous substitutions were calculated as described previously (61). Red, nonsynonymous substitutions; green, synonymous substitutions; blue horizontal bars, transmembrane domains; vertical bars at the bottom, substitution sites.
The origins of the divergence seen in PDR5 are unclear. The closest matching sequence to YJM789 PDR5 in GenBank is S288c PDR5. PDR15 is the closest paralog to PDR5 in both strains, yet there is >25% divergence at the protein level between PDR5 and PDR15 in each genome. Because this divergence is higher than the divergence observed for the two PDR5 orthologs (5.3%) (SI Fig. 8), ectopic recombination between PDR5 and PDR15 may not be the cause of high divergence between YJM789 and S288c. Interestingly, the corresponding gene products in S. paradoxus and, potentially, in RM11-1a are both truncated. The PDR5 gene product in YJM789 appears to be inactive for at least one substrate, resulting in cycloheximide hypersensitivity in this strain (25). No obvious loss-of-function mutations (frameshift or nonsense) were detected in the coding sequence, although such mutations might have been anticipated if there had been selection for loss of Pdr5p function or if there had been random genetic drift after inactivation.
Indels.
Indels between the genome sequences of YJM789 and S288c were identified by using chromosome-by-chromosome examinations of the sequence alignments to reveal the physical gaps (see Fig. 1 for indel analysis results for chromosome XIV and SI Fig. 5 for the other 15 chromosomes). Within the high-quality YJM789 sequence, 275,836 bp were identified in the S288c genome that are absent in YJM789, and 48,764 bp in the YJM789 genome that are absent in S288c. Furthermore, 269 indels are >100 bp, and 5,600 indels are <100 bp (see SI Fig. 9 for the distribution of indel size). The indels often involve Ty transposable elements. Assembling shotgun sequence reads for transposable elements is particularly challenging because of sequence similarities. Therefore, the identification of these elements in YJM789 is preliminary. We identified 17 Ty elements in the YJM789 genome, all of which are Ty1, Ty2, and Ty5, compared with 50 in S288c. Ty3 and Ty4 were not found and are suspected to be absent from the YJM789 genome, a result supported by the absence of hybridization to probes covering these genes during array analysis (26).
Gene Product Polymorphisms and Their Phenotypic Consequences.
Many orthologs between YJM789 and S288c contain nucleotide polymorphisms that affect either the sequence or length of the corresponding gene products. The 5% most variable genes with nonsynonymous polymorphisms (and no indels) are found to be significantly enriched in unknown functions (SI Fig. 10 for gene ontology category comparison). Gene product length polymorphisms resulting from in-frame or out-of-frame indels, ORF fusions, nonsense mutations (SNPs), and Ty polymorphisms (SI Data Sets 2 and 3 list selected ORFs of each category) are less abundant and likely to impact gene product functions.
There are cases where two or more ORFs annotated as separate in S288c appear to be a single ORF in YJM789 (SI Data Set 3). One case is NFT1, annotated in S288c as two genes (YKR103W and YKR104W). The stop codon becomes a tyrosine-encoding TAT codon in YJM789, as well as in several other Saccharomyces species, resulting in a longer ORF (44). Another case involves the S288c ORFs, YJL107C and PRM10, which appear to be a single ORF in YJM789 and other fungi (45, 46).
Although sequence information alone is inadequate for predicting the phenotypic consequence of polymorphisms, there are a few cases for which such consequences can be proposed. One example is the inactivating missense polymorphism found in the S288c AMN1 gene, which is responsible for yielding nonclumpy cells. RM11-1a cells do not separate efficiently, form clumps, and lack this polymorphism (46). Likewise, the YJM789 AMN1 gene contains the same SNP as that of RM11-1a. Although YJM789 is less clumpy than RM11-1a, it is much more clumpy than S288c (Gael Yvert, personal communication).
In addition, several genes with product length polymorphisms appear to be functional in YJM789 but apparently not functional in S288c (SI Data Set 3). For example, the S288c HAP1 gene contains a partially inactivating Ty1 insertion, resulting in a hap1 hypomorphic mutation (47), whereas the YJM789 HAP1 gene has no insertion. The S288c FLO8 gene (a transcription factor required for pseudohyphal formation) contains an inactivating amber mutation (48), yet YJM145 (a diploid isogenic with YJM789) forms abundant pseudohyphae (20), and its FLO8 ORF has no amber mutation.
Many genes bearing intragenic tandem repeats have different frame repeat numbers between S288c and YJM789. Several of these genes encode cell surface proteins (such as TIR1, HSP150, FIT1, AGA1, MNN4, and FLO10), and their variation in tandem repeats may generate functional cell surface variability that has been reported to contribute to a rapid adaptation to the environment and possibly host immune evasion (49).
Mitochondrial Genome.
The mitochondrial sequence of YJM789 is collinear with that of strain S288c and approximately the same size (86,214 vs. 85,779 bp). For much of the mitochondrial genome, sequence identity is >98%. Nevertheless, these genomes differ in several ways. Strain YJM789 is missing ≈54 GC-rich transposable elements (50) but contains 17 that are not present in S288c, none of which disrupt verified genes.
One particularly interesting gene present in the YJM789 mitochondrial genome is a maturase, an ortholog of Candida stellata cox-i2. In YJM789, this ortholog is encoded partly in an additional intron of COX1 (intron 4). In addition to the cox-i2 ortholog, three regions of high sequence divergence exist. Intron 6 of COX1, which is 1,487 bp, is approximately the same length as intron 5 of COX1 of S288c (1,365 bp) but essentially shows no sequence conservation. High variation is found between the ATP6 gene and tRNA-Glu and includes the region encoding the putative RF3 maturase protein. Furthermore, between COX2 and RNA-Phe is a 1,417-bp region, which encodes the hypothetical RF1 gene that has only 72% identity to the corresponding region of S288c.
Implications.
The YJM789 genome sequence is marked by extensive polymorphisms relative to the laboratory strain S288c throughout the nuclear and mitochondrial genomes. The ≈60,000 SNPs scattered over the genome alignment represent a SNP frequency of 1 in 164 bp (0.6%), which is higher than the divergence between human beings (0.1–0.01%). High SNP frequencies together with indels could account for the reduced spore viability seen in crosses of these two strains: 87.4% for YJM789/S288c background hybrids, compared with 97.6% for S288c/S288c (19). In comparison with SNPs, the number of indels measuring >100 bp (269) is moderate. Although the indels involve ≈324 kb (within the aligned 98% of the S288c genome), much of them represent repeat sequences, which could suggest that SNPs might be a primary cause of heritable phenotypic variation between these two strains.
Although the idea of horizontal gene transfer has been accepted in bacteria (51), eukaryotic genomes were initially thought to be units that do not exchange genetic information (52). The YJM789 genome provides preliminary evidence to suggest a putative horizontal transfer of YJM-GNAT from bacteria and a potential introgression of an ≈12-kb chromosome I sequence from closely related yeast. Although further analysis with sequences of more yeast strains will be informative for proof, the possibilities of such horizontal genetic exchanges are in line with an increasing number of reports describing introgression or horizontal genetic exchange in Saccharomyces sensu stricto species (36, 40, 53–55).
Finally, we made the YJM789 genome a free-to-access resource that marks an initial step toward a more complete set of reference sequences for the S. cerevisiae species. The benefits of complete genome information of several individuals can soon be explored. One key application will be the development of new technologies to interrogate the genome content of several S. cerevisiae strains by including, for example, polymorphism-specific probes on tiling microarrays. These technologies have promise to further advance applications in yeast to define the contribution of sequence variants to heritable traits. Importantly, applied to YJM789, these technologies will help us to understand how sequence polymorphisms change the information encoded in the genome to confer pathogenicity.
Materials and Methods
Gene Prediction and Comparison.
We used three different gene-prediction methods to identify potential ORFs: (i) directly mapping genes by using the S288c-verified ORFs from the Saccharomyces database (56), (ii) ORF calling based on the positions of start and stop codons, and (iii) GLIMMERM (57). Ortholog assignments were required to meet all of the following criteria: reciprocal best match with an E value of the high-scoring segment pairs of ≤1 × 10−5, an identity of ≥40%, and a match length of at least 75% of both protein lengths. Homologs were identified in cases for which no reciprocal best hit was obtained as the nearest S288c best hit homolog, with threshold requirements as described above. YJM789 genes without S288c homologs were defined as YJM789-specific.
ORF Annotation.
The gene names, functional descriptions, and gene ontology categories for the YJM789 genes with S288c orthologs or homologs were annotated according to their S288c counterparts. The annotation of specific genes of YJM789 was based on comparison to the nonredundant database. For the YJM789 genes with S288c homologs that contained frameshift, indel, or missense mutations, the nature of the potentially inactivating polymorphism was identified. Complete annotations are provided in SI Data Set 1.
Genome Alignment.
The public software BLASTN (58) and MUMmer (59) were used to align the sequences of the high-quality contigs of YJM789 to the individual S288c chromosome sequences. The results of these two analyses were checked manually and combined. In regions of disagreement between the alignment programs, the alignment with highest sequence similarity was chosen. The polymorphism sets were derived from the final chromosome alignment of S288c with the 31 YJM789 contigs of at least 10 kb in length. Insertion and deletion events were parsed by counting the number and size of the alignment gaps between S288c and YJM789 contigs. SNPs were detected by base substitution in the alignment, provided that the bases of the YJM789 sequence had a quality score of at least 40 (60). All polymorphisms in repeat sequences were excluded.
Additional Materials and Methods.
Further details are found in SI Text.
Supplementary Material
Acknowledgments
We thank Gael Yvert for information regarding YJM789 clumpiness and Michael Knop, Rui Wang-Sattler, Guohui Ding, Kang Tu, Lin Tao, Hao Xu, Hong Yu, and Ziliang Qian for helpful discussion, suggestions, and assistance with calculations. The RM11-1a sequence was obtained from the S. cerevisiae RM11-1a Sequencing Project, Broad Institute of Harvard and Massachusetts Institute of Technology. This work was supported by National Institutes of Health Grants HG02052 (to R.W.D.), GM068717 (to R.W.D. and L.M.S.), and HG000205 (to R.W.D. and L.M.S.); China National Basic Key Research Program Grants 2003CB715901 (to Y.L.), 2004CB518606 (to Y.L.), 2006CB910700 (to Y.L.), 2004CB720103 (to Z.C.), and 2006AA02Z317 (to Z.C.); China National Natural Science Foundation Grants 30500107 and 30670953 (to Z.C.); and Science and Technology Commission of Shanghai Municipality Grant 06PJ14072 (to Z.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Whole Genome Shotgun has been deposited in the GenBank database (project accession no. AAFW00000000). The version described in this article is the second version (accession no. AAFW02000000). The accession no. for the mitochondrial genome is EU004203.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701291104/DC1.
References
- 1.Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, Davis RW. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 2.Brem RB, Yvert G, Clinton R, Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 3.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 4.Deutschbauer AM, Davis RW. Nat Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Ari G, Zenvirth D, Sherman A, David L, Klutstein M, Lavi U, Hillel J, Simchen G. PLoS Genet. 2006;2:e195. doi: 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perlstein EO, Ruderfer DM, Ramachandran G, Haggarty SJ, Kruglyak L, Schreiber SL. Chem Biol. 2006;13:319–327. doi: 10.1016/j.chembiol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Sinha H, Nicholson BP, Steinmetz LM, McCusker JH. PLoS Genet. 2006;2:e13. doi: 10.1371/journal.pgen.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 9.Kumar A, Snyder M. Nat Rev Genet. 2001;2:302–312. doi: 10.1038/35066084. [DOI] [PubMed] [Google Scholar]
- 10.Steinmetz LM, Davis RW. Nat Rev Genet. 2004;5:190–201. doi: 10.1038/nrg1293. [DOI] [PubMed] [Google Scholar]
- 11.Mortimer RK, Johnston JR. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu Z, David L, Petrov D, Jones T, Davis RW, Steinmetz LM. Proc Natl Acad Sci USA. 2005;102:1092–1097. doi: 10.1073/pnas.0409159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronald J, Tang H, Brem RB. Genetics. 2006;174:541–544. doi: 10.1534/genetics.106.060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enache-Angoulvant A, Hennequin C. Clin Infect Dis. 2005;41:1559–1568. doi: 10.1086/497832. [DOI] [PubMed] [Google Scholar]
- 15.Murphy AR, Kavanagh KA. Med Mycol. 2001;39:123–127. doi: 10.1080/mmy.39.1.123.127. [DOI] [PubMed] [Google Scholar]
- 16.Ponton J, Ruchel R, Clemons KV, Coleman DC, Grillot R, Guarro J, Aldebert D, Ambroise-Thomas P, Cano J, Carrillo-Munoz AJ, et al. Med Mycol. 2000;38(Suppl 1):225–236. doi: 10.1080/mmy.38.s1.225.236. [DOI] [PubMed] [Google Scholar]
- 17.de Llanos R, Querol A, Peman J, Gobernado M, Fernandez-Espinar MT. Int J Food Microbiol. 2006;110:286–290. doi: 10.1016/j.ijfoodmicro.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 18.Tawfik OW, Papasian CJ, Dixon AY, Potter LM. J Clin Microbiol. 1989;27:1689–1691. doi: 10.1128/jcm.27.7.1689-1691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCusker JH, Clemons KV, Stevens DA, Davis RW. Genetics. 1994;136:1261–1269. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCusker JH, Clemons KV, Stevens DA, Davis RW. Infect Immun. 1994;62:5447–5455. doi: 10.1128/iai.62.12.5447-5455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byron JK, Clemons KV, McCusker JH, Davis RW, Stevens DA. Infect Immun. 1995;63:478–485. doi: 10.1128/iai.63.2.478-485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clemons KV, McCusker JH, Davis RW, Stevens DA. J Infect Dis. 1994;169:859–867. doi: 10.1093/infdis/169.4.859. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein AL, McCusker JH. Genetics. 2001;159:499–513. doi: 10.1093/genetics/159.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingsbury JM, Goldstein AL, McCusker JH. Eukaryot Cell. 2006;5:816–824. doi: 10.1128/EC.5.5.816-824.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, Davis RW. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 26.Winzeler EA, Lee B, McCusker JH, Davis RW. Parasitology. 1999;118:S73–S80. doi: 10.1017/s0031182099004047. [DOI] [PubMed] [Google Scholar]
- 27.Winzeler EA, Castillo-Davis CI, Oshiro G, Liang D, Richards DR, Zhou Y, Hartl DL. Genetics. 2003;163:79–89. doi: 10.1093/genetics/163.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, Kruglyak L. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- 29.Goto K, Iwatuki Y, Kitano K, Obata T, Hara S. Agr Biol Chem. 1990;54:979–984. [PubMed] [Google Scholar]
- 30.Ness F, Aigle M. Genetics. 1995;140:945–956. doi: 10.1093/genetics/140.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. Antimicrob Agents Chemother. 1993;37:1896–1903. doi: 10.1128/aac.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Draker KA, Wright GD. Biochemistry. 2004;43:446–454. doi: 10.1021/bi035667n. [DOI] [PubMed] [Google Scholar]
- 33.Vetting MW, Magnet S, Nieves E, Roderick SL, Blanchard JS. Chem Biol. 2004;11:565–573. doi: 10.1016/j.chembiol.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 34.Salzberg SL, White O, Peterson J, Eisen JA. Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 35.Török T, Mortimer RK, Romano P, Suzzi G, Polsinelli M. J Ind Microbiol Biotechnol. 1996;17:303–313. [Google Scholar]
- 36.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 37.Poirey R, Despons L, Leh V, Lafuente MJ, Potier S, Souciet JL, Jauniaux JC. Microbiology. 2002;148:2111–2123. doi: 10.1099/00221287-148-7-2111. [DOI] [PubMed] [Google Scholar]
- 38.Leh-Louis V, Wirth B, Potier S, Souciet JL, Despons L. Genetics. 2004;167:1611–1619. doi: 10.1534/genetics.104.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sniegowski PD, Dombrowski PG, Fingerman E. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 40.Liti G, Louis EJ. Annu Rev Microbiol. 2005;59:135–153. doi: 10.1146/annurev.micro.59.030804.121400. [DOI] [PubMed] [Google Scholar]
- 41.Naumov GI. Mol Gen Mikrobiol Virusol. 1987;2:3–7. [PubMed] [Google Scholar]
- 42.Naumov GI, Naumova ES, Lantto RA, Louis EJ, Korhola M. Yeast. 1992;8:599–612. doi: 10.1002/yea.320080804. [DOI] [PubMed] [Google Scholar]
- 43.Hunter N, Chambers SR, Louis EJ, Borts RH. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 44.Mason DL, Mallampalli MP, Huyer G, Michaelis S. Eukaryot Cell. 2003;2:588–598. doi: 10.1128/EC.2.3.588-598.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brachat S, Dietrich FS, Voegeli S, Zhang Z, Stuart L, Lerch A, Gates K, Gaffney T, Philippsen P. Genome Biol. 2003;4:R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sychrova H, Braun V, Potier S, Souciet JL. Yeast. 2000;16:1377–1385. doi: 10.1002/1097-0061(200011)16:15<1377::AID-YEA637>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 47.Gaisne M, Becam AM, Verdiere J, Herbert CJ. Curr Genet. 1999;36:195–200. doi: 10.1007/s002940050490. [DOI] [PubMed] [Google Scholar]
- 48.Liu H, Styles CA, Fink GR. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Nat Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Séraphin B, Simon M, Faye G. Nucleic Acids Res. 1985;13:3005–3014. doi: 10.1093/nar/13.8.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gogarten JP, Townsend JP. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 52.Mayr E. Systematics and the Origin of Species. New York: Columbia Univ Press; 1942. [Google Scholar]
- 53.Liti G, Barton DB, Louis EJ. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dujon B. Curr Opin Genet Dev. 2005;15:614–620. doi: 10.1016/j.gde.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Hall C, Brachat S, Dietrich FS. Eukaryot Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirschman JE, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hong EL, Livstone MS, Nash R, et al. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Nucl Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 61.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.