Abstract
APOBEC3G (A3G) is a single-stranded DNA cytidine deaminase that targets retroviral minus-strand DNA and has potent antiviral activity against diverse retroviruses. However, the mechanisms of A3G antiviral functions are incompletely understood. Here we demonstrate that A3G, A3F, and, to a lesser extent, the noncatalytic A3GC291S block human immunodeficiency virus type 1 (HIV-1) replication by interfering with proviral DNA formation. In HIV-1 virions, A3G interacted with HIV-1 integrase and nucleocapsid, key viral factors for reverse transcription and integration. Unlike A3G, the weak antiviral A3C cytidine deaminase did not interact with either of these factors and did not affect viral reverse transcription or proviral DNA formation. Thus, multiple steps of the HIV-1 replication cycle, most noticeably the formation of proviral DNA, are inhibited by both cytidine deamination-dependent and -independent mechanisms.
Human cytidine deaminase APOBEC3G (A3G) was the first APOBEC protein to be identified as a potent inhibitor of Vif-deficient human immunodeficiency virus type 1 (HIV-1) (48). In order to successfully replicate in their hosts, lentiviruses such as HIV-1 and simian immunodeficiency virus (SIV) encode the Vif protein, which induces polyubiquitination and degradation of multiple APOBEC3 molecules (12, 30, 31, 38, 39, 49, 53, 61). Vif molecules of HIV-1 and SIV are newly identified substrate receptor proteins that assemble with Cul5, ElonginB, ElonginC, and Rbx1 to form an E3 ubiquitin ligase (22, 30, 34, 39, 61, 62). Several human APOBEC3 proteins, including A3G, APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), and APOBEC3F (A3F), as well as APOBEC3 from the mouse, have recently been shown to act as broad antiviral factors against HIV-1, SIV, murine leukemia virus (MuLV), endogenous retroelements, and hepatitis B virus (4-6, 9, 13, 15, 16, 18, 24, 25, 29, 32, 35-37, 40, 42, 44, 47, 51, 54-56, 58-60, 64, 65).
The mechanisms that undergird the antiviral functions of A3G are not yet fully understood. It is clear that virion packaging of A3G is critical for its antiviral function, and this packaging is mediated through the nucleocapsid (NC) domain of HIV-1 Gag molecules in the absence of Vif (1, 14, 46, 63). In newly infected cells, A3G induces C-to-U mutations in the minus-strand viral DNA during reverse transcription, resulting in G-to-A mutations in the coding strand (18, 25, 36, 37, 54, 60, 64). Successful packaging of active cytidine deaminases into virions does not, however, guarantee antiviral action or efficient deamination of viral DNA. Both A3C and A3G are robust enzymes that generate mutations in Escherichia coli DNA (19). However, although A3G can drastically reduce HIV-1 infectivity, A3C is relatively ineffective (4, 24, 58, 59, 65). Furthermore, A3C induces significantly fewer G-to-A mutations in HIV-1 DNA than does A3G (59). Therefore, there appear to be additional determinants that intervene between the packaging of cytidine deaminases into retroviral particles and the manifestation of antiviral activity.
Cytidine deamination may not be the only antiviral function of A3G. A3G mutants that lack cytidine deamination activity still demonstrate substantial anti-HIV-1 activity (41, 50), and non-deamination-mediated inhibition of HIV-1 reverse transcription by APOBEC3 proteins has been reported recently (3, 17). A3G also mediates weak antiviral activity against human T-cell leukemia type 1 without causing cytidine deamination of viral DNA and does not require either the N-terminal or the C-terminal active sites of the enzyme (45). Low-molecular-mass A3G restricts HIV-1 infection in resting CD4+ T cells without inducing significant cytidine deamination of HIV-1 DNA (11). In addition, cytidine deaminase mutants of A3G have been shown to suppress hepatitis B virus (55).
In the present study, we demonstrate that noncatalytic A3GC291S as well as A3G can block HIV-1 replication, mainly through interference with reverse transcription and viral DNA integration. In HIV-1 virions, A3G interacted with HIV-1 integrase (IN) and NC, which are known to be important for reverse transcription and integration. The C-terminal domain of HIV-1 IN and the first linker region of A3G were found to be required for IN and A3G interaction. On the other hand, the weak antiviral A3C did not interact with IN or NC and had little effect on reverse transcription and integration. Furthermore, A3G, but not A3GC291S, affected the nuclear accumulation of viral 2-long terminal repeat (2-LTR) circle DNA.
MATERIALS AND METHODS
Plasmids.
The A3G-HA and HIV-1ΔVif expression vectors have been described previously (61). A3G truncation constructs have been previously described (8). pHCMV-VSV-G was obtained from the NIH AIDS Research Reference and Reagent Program. The A3C-HA expression vector was generously provided by M. Malim. A3GC291S and A3GC291S-HA were constructed by the PCR-based mutagenesis method using the following primers and pA3G-HA as template: forward, GCTCTTCAGCAGCGCACAGGAAATGGCTAA; reverse, TTAGCCATTTCCTGTGCGCTGCTGAAGAGC. The C-terminal myc-tagged integrase construct was amplified from the codon-optimized HIV-1 Pol gene (23) using the following primers: IN 5′ SalI primer, 5′-GTCGACACCATGTTCCTGGACGGCATCGAC; IN 3′ BamHI/Myc primer, 5′-GGATCCTCAAAGATCTTCTTCTGATATGAGTTTTTGTTCGTCCTCATCCTGC. The PCR products were digested with SalI and BamHI and cloned into VR1012 to generate pINmyc. The pINΔNTD, pINΔCTD, pINΔCCD1, and pΔCCD2 mutant constructs were constructed by the PCR-based mutagenesis method using the follow primers: pINΔNTD forward, 5′-AGTCGACCATGCACGGCCAGGTGGACTG; reverse, 5′-GGATCCTCAAAGATCTTCTTCTGATATGAGTTTTTGTTCGTCCTCATCCTGC; pINΔCTD forward, 5′-GAACAAAAACTCATATCAGAAGAAG; reverse, 5′-CTCCCGGGTCTGGATGTC; pINΔCCD1 forward, 5′-GCCGGCATCCAGCA; reverse, 5′-GCCGTGCATGGCCTC; pINΔCCD2 forward, 5′-CTGCAGAAGCAGATCATTAAG; reverse, 5′-CCACCAGCAGGCTGC.
Antibodies and cells.
293T cells and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) with 10% fetal bovine serum (FBS) and gentamicin (5 μg/ml) (D-10 medium) and passaged upon confluence. The following antibodies were used for this study: anti-hemagglutinin (HA) antibody-agarose conjugate (catalog no. 1815016; Roche), an HIV-1-positive serum, goat anti-p7 antiserum, anti-HA mouse monoclonal antibody (MAb; catalog no. MMS-101R-10000; Covance), and anti-human ribosomal P antigen antibody (catalog no. HP0-0100; Immunovision). Anti-HIV-1 integrase and anti-human A3G were obtained from the NIH AIDS Research Reference and Reagent Program (catalog nos. 757, 758, and 9906). HIV-1 p24 protein was detected using the HIV-1 p24 enzyme-linked immunosorbent assay kit (Perkin-Elmer).
Transfection, infection, detection of integrated viral DNA, and virus purification.
DNA transfection was carried out using Lipofectamine 2000 (Invitrogen) as described by the manufacturer. Virion-associated viral proteins were prepared from cell culture supernatants and separated from cellular debris by centrifugation at 3,000 rpm for 30 min in a Sorvall RT 6000B centrifuge and filtration through a 0.2-μm-pore-size membrane. Virus particles were concentrated by centrifugation through a 30% sucrose cushion at 100,000 × g for 2 h in a Sorvall Ultra80 ultracentrifuge.
For single-round virus infection, 293T cells were cotransfected with an HIV-1 vector (HXB2NeoΔVif) and vesicular stomatitis virus (VSV)-G plus various APOBEC3 expression vectors or a control vector (pcDNA3.1). Culture media were removed 6 h after transfection and washed twice with DMEM containing 1% FBS. Cells were cultured in fresh medium for another 60 h. Supernatants containing viruses were treated with DNase I (100 U/ml) at 37°C for 1 h and used to infect HeLa cells. Infected cells were harvested at the indicated time points, washed three times with phosphate-buffered saline (PBS), and treated with DNase I (100 U/ml) at 37°C for 1 h. The cells were washed again with PBS and were lysed with lysis buffer (QIAGEN) at 70°C for 10 min. DNA was isolated from the cell lysates using a DNeasy DNA isolation kit (QIAGEN). DNA samples were further treated with DpnI at 37°C for 16 h to remove any potential plasmid contamination prior to PCR amplification.
To study gene expression from integrated HIV-1 DNA, infected HeLa cells were cultured in the presence of G418 (1 mg/ml). Neomycin-resistant cell clones were counted 3 weeks after infection.
Immunoblot analysis.
Cells were collected 48 h after transfection. Cell and viral lysates were prepared as previously described (61). Cells (1 × 105) were lysed in 1× loading buffer (0.08 M Tris, pH 6.8, with 2.0% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue). The samples were boiled for 10 min, and proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred onto nitrocellulose membranes and probed with various primary antibodies against the indicated proteins. Secondary antibodies were alkaline phosphatase-conjugated anti-human, anti-rabbit, anti-mouse, or anti-goat immunoglobulin G (Jackson Immunoresearch, Inc.), and staining was carried out with 5-bromo-4-chloro-3-indolylphosphate and nitro blue tetrazolium solutions prepared from chemicals obtained from Sigma.
Immunoprecipitation.
For immunoprecipitation experiments, the protein-expressing cells were harvested and washed twice with cold PBS and lysed with PBS containing 0.5% Triton X-100 and protease inhibitor cocktail (Roche) at 4°C for 1 h. Cell lysates were clarified by centrifugation at 10,000 × g for 30 min at 4°C. For virion immunoprecipitation, virion-associated viral proteins were prepared from cell culture supernatants and separated as described above and then lysed with PBS containing 0.5% Triton X-100 and protease inhibitor cocktail (Roche) at 4°C for 1 h. Anti-HA agarose (Roche) or anti-myc antibody (Upstate) and protein G (Invitrogen) were mixed with precleared cell lysates or virion lysates and incubated at 4°C for 3 h. The reaction mixture was then washed three times with cold PBS and eluted with 0.1 M glycine-HCl buffer, pH 2.0. The eluted materials were subsequently analyzed by SDS-PAGE and immunoblotting. The eluted materials were subsequently analyzed by SDS-PAGE and immunoblotting.
Hypermutation analysis.
293T cells were cotransfected with the indicated HIV-1 expression vectors and VSV-G plus the indicated A3G expression vector or control vector pcDNA3.1. Culture supernatants were collected at 72 h after transfection and treated with DNase I (100 U/ml) at 37°C for 1 h. HeLa cells (1 × 106) were infected with DNase I-treated HIV-1 (150 ng p24 equivalent) for 4 h and then washed with fresh medium. Infections were carried out for another 12 h, and infected cells were washed three times with PBS and treated with DNase I (100 U/ml) at 37°C for 1 h. The cells were again washed with PBS and then lysed in lysis buffer at 70°C for 1 h. DNA was isolated from lysates of infected cells using a DNeasy DNA isolation kit (QIAGEN) and treated with DpnI at 37°C for 16 h to remove any potential plasmid contamination prior to PCR amplification. A 650-bp DNA fragment covering a portion of nef, U3, and R of HIV-1 was amplified with Hotstar Taq DNA polymerase (QIAGEN) using the primers HIV-1-F, 5′-AGGCAGCTGTAGATATTAGCCAC, and HIV-1-R, 5′-GTATGAGGGATCTCTAGCTACCA. The PCR products were cloned into the TOPO TA-cloning vector pCR4 (Invitrogen). The nucleotide sequences of 15 clones from each infected culture sample were determined and analyzed using Hypermut software (43).
qRT-PCR.
Virus infection and DNA extraction were performed as described above. DNA was used as templates for real-time PCR. The primers for the detection of early reverse transcription (ERT), late reverse transcription (LRT), 2-LTR circle DNA, and Alu-LTR-based integrated DNA have been described already (7). For ERT, LRT, and 2-LTR DNA, 300 nM forward primer, 300 nM reverse primer, 100 nM probe, and 100 to 500 ng of template DNA were used in a 30-μl reaction volume. After initial incubations at 50°C for 2 min and 95°C for 10 min, 40 cycles of amplification were carried out for 15 s at 95°C, followed by 1 min at 60°C. Integration assays used 50 nM forward primer, 900 nM reverse primer, 100 nM probe primer, and 250 to 500 ng of template DNA in a 50-μl volume. PCR cycling began with a denaturation step (95°C for 10 min), followed by 45 cycles of amplification (95°C for 15 s, 60°C for 1 min, and 72°C for 1 min). Reactions were analyzed in TaqMan universal PCR mix (Applied Biosystems) on an ABI prism 7000 (Applied Biosystems). Quantitative real-time PCR (qRT-PCR) primers/probes were tested on serially diluted plasmids or purified DNA from HIV-1-infected cells containing the target sequence. Only threshold cycle values within the linear range of detection were used to calculate DNA levels in experimental samples.
RESULTS
Noncatalytic A3GC291S inhibits HIV-1 reverse transcription and integration.
A recent study found that the antiviral function of A3G can be dissociated from its cytidine deaminase activity (41). A single-amino-acid substitution in the C-terminal catalytic domain (C291S) of A3G abolished its cytidine deamination activity in E. coli (41) as well as its ability to induce cytidine deamination of HIV-1 DNA during infection (Fig. 1A). To examine the effect of the noncatalytic A3GC291S on HIV-1 replication, we utilized well-established qRT-PCR assays (7) to monitor the synthesis of early (ERT) and late (LRT) products of viral reverse transcription and the formation of viral 2-LTR circle DNA (an indication of nuclear import of viral DNA) and integrated viral DNA (Fig. 1B). For this purpose, we cotransfected 293T cells with HIV-1ΔVif (NL4-3ΔVif), an expression vector for VSV-G, and the expression vector for A3GC291S or a control vector. At 72 h after transfection, the virus-containing supernatants were used to infect HeLa cells. The transient expression of VSV-G proteins allowed the viruses to infect HeLa cells only once, so that the effects of A3GC291S on HIV-1 replication could be analyzed in a single round of infection. At various time points following infection, infected cells were harvested and various viral DNA products were examined by qRT-PCR using specific primers/probes.
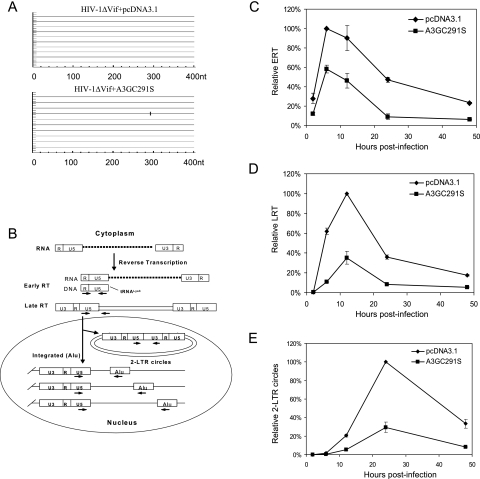
FIG. 1.
Effects of A3GC291S on HIV-1 reverse transcription and nuclear import during a single-round infection of HeLa cells. HIV-1ΔVif was cotransfected with the pcDNA3.1 or A3GC291S expression vector into 293T cells along with the VSV-G expression vector. At 72 h posttransfection, viruses were collected and used to infect HeLa cells. The infected cells were harvested at different time points. (A) A3GC291S is defective for cytidine deamination of HIV-1 DNA. The proviral DNA was extracted at 16 h after infection, and fragments of HIV-1 in the region of the Nef-U5 from virus-infected cultures in the presence of A3GC291S or a control vector were amplified, cloned, and sequenced. G-to-A mutations are shown. (B) Schematic of the early events in HIV-1 replication and the locations of PCR primers. HIV-1 genomic RNAs were reverse transcribed to yield linear viral DNAs. Of these, some were degraded, some were circularized to 1- or 2-LTR circles, and some were integrated. Arrows indicate the approximate locations of the PCR primers used to detect the different HIV-1 DNA forms. Real-time PCR was used for time course analysis of HIV-1 ERT (C), LRT (D), and 2-LTR circle DNA formation (E). nt, nucleotide.
ERT products were detectable at 2 h after infection, and their levels peaked at 6 h after infection in both the presence and absence of A3GC291S (Fig. 1C). However, A3GC291S was associated with a reduced accumulation of ERT at all time points after infection (Fig. 1C). At 6 h, the ERT levels produced by A3GC291S-transfected cells were about 42% lower than those from control virus-infected cells (Fig. 1C). The LRT products were first detected at 6 h after infection and peaked at 12 h after infection in both the presence and the absence of A3GC291S (Fig. 1D). Transfection of A3GC291S reduced the accumulation of LRT at 6 h after infection and decreased the LRT peak level at 12 h after infection by about 62% compared to the control virus infection lacking A3GC291S (Fig. 1D). A3GC291S is defective for cytidine deamination, and therefore it would not be expected to trigger viral DNA degradation by uracil DNA glycosylase/apurinic-apyrimidinic endonuclease. There was no evidence that ERT or LRT products disappeared more quickly in the presence of A3GC291S than in its absence. Thus, the noncatalytic A3GC291S was likely inhibiting viral DNA synthesis during reverse transcription.
To investigate the effect of A3GC291S on events beyond reverse transcription, we also monitored the synthesis of 2-LTR circle DNA in the presence or the absence of A3GC291S. Synthesis of 2-LTR circle DNA peaked at 24 h after infection, and A3GC291S reduced the accumulation of 2-LTR DNA by about 65% compared to the control virus lacking A3GC291S (Fig. 1E); this reduction was similar in magnitude to that seen for LRT (Fig. 1D). The production of viral 2-LTR circle DNA is an indication of productive nuclear entry by viral DNA (52). These results indicate that A3GC291S did not affect the entry of the viral DNA into the nucleus, a process that is required for the formation of 2-LTR circle DNA.
To monitor the formation of provirus, we analyzed integrated HIV-1 DNA levels in virus-infected HeLa cell samples by means of a quantitative assay using Alu-LTR-based qRT-PCR (Fig. 1B), as adapted from the method described by Butler et al. (7). The quantity of integrated HIV-1 DNA in cell samples infected with HIV-1ΔVif in the presence of A3GC291S was approximately 10% of that in control HIV-1ΔVif-infected cells lacking A3GC291S (Fig. 2A). Since the LRT level in cells cotransfected with HIV-1ΔVif in the presence of A3GC291S was approximately 38% of that in control HIV-1ΔVif-infected cells in the absence of A3GC291S (Fig. 1D) and 2-LTR circle viral DNA levels in cells infected with HIV-1ΔVif and A3GC291S were about 35% of those in control cells (Fig. 1E), the 3-fold reduction in viral reverse transcription/nuclear entry produced by A3GC291S cannot fully account for the 10-fold reduction we observed in the amount of integrated viral DNA. Thus, HIV-1 integration was apparently inhibited in the presence of A3GC291S (by approximately threefold).
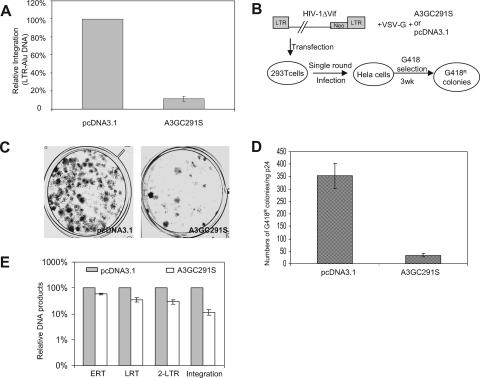
FIG. 2.
Effect of A3GC291S on HIV-1 integration. (A) Analysis of HIV-1 DNA integration in HeLa cells by real-time PCR. DNA samples were prepared as described for Fig. 1. The location of primers is indicated in Fig. 1B. (B) Schematic representation of the experimental procedure for stable gene expression after HIV-1 integration. (C) A set of plates stained with Giemsa 3 weeks (3wk) after G418 selection. (D) Quantification of the number of G418-resistant colonies, after selection in G418-containing medium, of HeLa cells infected with HIV-1ΔVif produced from 293T cells with VSV-G-pseudotyped HIV-1ΔVif virus in the presence of pcDNA3.1 or A3GC291S. (E) Comparison of ERT, LRT, 2-LTR, and integration (Alu-based qRT-PCR) in the absence or presence of A3GC291S at the peak points.
The Alu-LTR qRT-PCR data were also consistent with the results of a functional assay that requires HIV-1 integration. After viral DNA synthesis and entry into the nucleus, integration of viral DNA into the host genome is critical for the stable expression of viral genes. Therefore, we used an HIV-1ΔVif construct containing a G418-resistant gene in the region of nef (26) to generate viruses in the presence or absence of A3GC291S, along with an expression vector for VSV-G (Fig. 2B). Viruses were used to infect HeLa cells (only one round of infection), and the numbers of G418-resistant colonies were determined 3 weeks after G418 selection. A 10-fold-lower number of G418-resistant colonies was observed in the presence of A3GC291S than in its absence (Fig. 2C and D). Since nuclear viral DNA was reduced by only about 3-fold (Fig. 1E) and the stable expression of a marker gene in HIV-1 genome was reduced by 10-fold in the presence of A3GC291S, these data again indicate that A3GC291S blocks the integration of HIV-1 DNA.
A3G has multiple effects on HIV-1 replication.
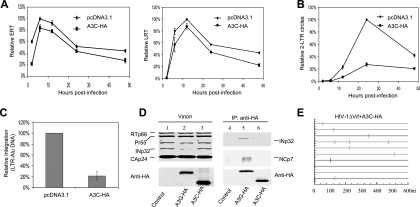
To determine whether catalytically active A3G affects similar steps in HIV-1 replication, we generated HIV-1ΔVif viruses from 293 or 293/A3G-HA cells (61). The expression level of A3G-HA in 293/A3G-HA cells (Fig. 3A, lane 1) was 2.5-fold higher than that of endogenous A3G in CD4+ H9 T cells (lane 2). In agreement with the data generated with A3GC291S, reverse transcription was reduced in the presence of A3G (Fig. 3B and C). A3G reduced the LRT level by about 60% at 12 h after infection (Fig. 3C). It is worth noting that A3G-mediated inhibition of plus-strand viral DNA (LRT) accumulation was observed at all the time points measured, beginning at 6 h after infection.
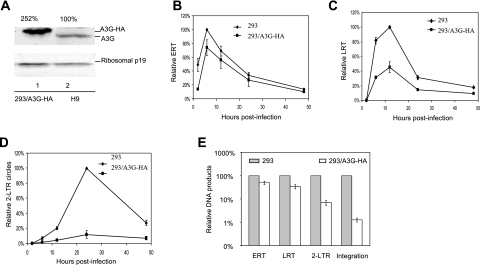
FIG. 3.
Effects of A3G on the early stages of HIV-1 replication during a single-round infection of HeLa cells. (A) Cell lysates of 293 cells stably expressing A3G-HA and H9 cells were used in an immunoblot assay to detect the expression of A3G using anti-APOBEC3G antibody. Ribosome p19 was used as a loading control. (B to E) 293 cells or 293 cells stably expressing A3G were transfected with HIV-1ΔVif-based vector and VSV-G. At 3 days posttransfection, HeLa cells were infected with HIV-1ΔVif that had been pretreated with DNase I. Total DNA was isolated from the infected cells at the indicated time points following infection, and early RT, late RT, and 2-LTR circles were quantified. Virus input was normalized according to the level of p24. The highest value at peak productivity of HIV-1 from 293T cells cotransfected with pcDNA3.1 was set to 100%. (B) Quantification of early RT products. (C) Quantification of late RT products. (D) Quantification of 2-LTR circles. (E) Comparison of ERT, LRT, 2-LTR, and integration (Alu-based qRT-PCR) in the absence or presence of A3G-HA at the peak points.
Unlike A3GC291S, A3G also caused a decrease in the detection of 2-LTR circle viral DNA after infection (Fig. 3D). In the presence of A3G, the decrease in 2-LTR circle DNA was greater than the decrease in LRT (Fig. 3E). When normalized to the LRT product levels, the production of 2-LTR circle DNA in the presence of A3G was approximately fourfold lower than that in its absence (Fig. 3E). Since the reduction in 2-LTR circle DNA was observed at multiple time points after infection, these findings suggest that we were seeing inhibition of the formation of 2-LTR circle DNA but not a rapid disappearance of this DNA. This reduction in 2-LTR circle DNA was not observed with the catalytic mutant A3GC291S (Fig. 1E).
Viruses produced from 293/A3G-HA cells showed a reduction of approximately 100-fold in proviral DNA formation, as measured by the Alu-based assay, compared to that generated from 293 cells that did not express detectable A3G (Fig. 3E). This difference could not be explained by the reduction in reverse transcription (2.5-fold difference) or the nuclear accumulation of 2-LTR DNA (4-fold difference). When normalized to 2-LTR DNA, the Alu-LTR-based qRT-PCR indicated a 10-fold-greater reduction in the presence of A3G-HA than in its absence. Thus, the formation of proviral DNA is a major target of A3G.
A3G and A3GC291S interact specifically with HIV-1 INp32 and NCp7 in released virions.
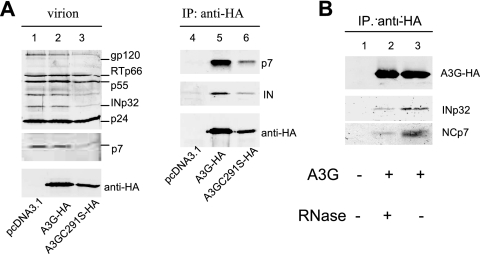
Cytidine deamination-defective A3GC291S is still able to inhibit reverse transcription and integration, suggesting that A3G may interact with viral protein(s) to mediate antiviral function. To examine whether A3G or A3GC291S interacts with viral protein(s) in HIV-1 virions, HIV-1ΔVif virions were produced in the presence or absence of A3G-HA or A3GC291S-HA from transfected 293T cells. Purified virions were analyzed by immunoblotting using pooled HIV-1-positive human sera. Various viral proteins, including Pr55Gag, Pr41Gag, CAp24, MAp17, RTp66, and INp32, were detected in virions lacking (Fig. 4A, lane 1) or containing A3G-HA (lane 2) or A3GC291S-HA (lane 3). Viral lysates were immunoprecipitated with an anti-HA antibody conjugated to agarose beads, and A3G-HA- or A3GC291S-HA-coimmunoprecipitated proteins were analyzed by immunoblotting using specific antibodies against the various HIV-1 proteins (Fig. 4A). INp32 and NCp7 were detected in the A3G-HA- and A3GC291S-HA-coimmunoprecipitated samples along with A3G-HA or A3GC291S-HA (Fig. 4A, lanes 5 and 6). They were not detected in viral lysates lacking A3G-HA or A3GC291S-HA (Fig. 4A, lane 4), indicating that INp32 or NCp7 did not nonspecifically bind to the anti-HA antibody-conjugated agarose beads. Interaction of A3G-HA or A3GC291S-HA with other viral proteins such as CAp24 was not detected (data not shown). Interaction between A3G-HA and INp32 in HIV-1 virions was reduced by RNase treatment (Fig. 4B), indicating a role for RNA in mediating the interaction between A3G and IN.
FIG. 4.
A3G and A3GC291S interact with HIV-1 INp32 and NCp7 in released virions. (A) HIV-1ΔVif was cotransfected with the pcDNA3.1, A3G-HA, or A3GC291S-HA expression vector into 293T cells. At 48 h posttransfection, viruses were collected and purified from culture supernatants. Viral lysates were analyzed by immunoblotting using pooled HIV-1-positive human sera for the detection of viral proteins or an anti-HA MAb for the detection of virion-packaged A3G-HA or A3GC291S-HA (A, lanes 1 to 3). Viral lysates were also immunoprecipitated with the anti-HA MAb conjugated to agarose beads, and coprecipitated samples were then analyzed by immunoblotting using the anti-INp32 polyclonal serum and the anti-NCp7 MAb to detect coimmunoprecipitated viral proteins or the anti-HA MAb to detect immunoprecipitated (IP) A3G-HA (A, lanes 4 to 6). (B) Interaction of IN with A3G in HIV-1 virions is RNA dependent. 293T cells were cotransfected with HIV-1ΔVif plus a control vector (lane 1) or an expression vector for A3G-HA (lanes 2 and 3). At 48 h posttransfection, viruses were harvested and viral lysates, either untreated (lanes 1 and 3) or treated with RNase (lane 2), were immunoprecipitated with the anti-HA MAb conjugated to agarose beads. Coprecipitated samples were then analyzed by immunoblotting using the anti-NCp7 (lower panel), anti-IN MAb (middle panel), or anti-HA MAb (upper panel).
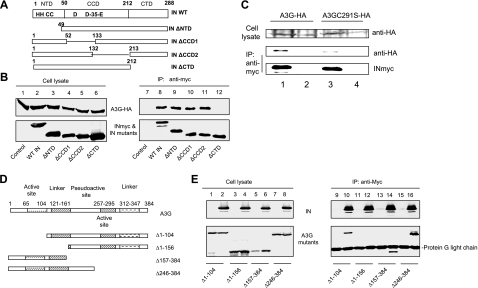
We have previously shown that the N-terminal region of NCp7 plays an important role in mediating its interaction with A3G (33). In order to map the regions in HIV-1 IN that mediate A3G binding, we constructed a series of mutant HIV-1 IN expression vectors. HIV-1 IN contains three well-defined structural domains: an N-terminal domain (NTD) that includes an HHCC zinc-binding motif, a central catalytic core domain (CCD), and a C-terminal domain (CTD). Four in-frame deletion mutants of HIV-1 IN constructs containing an NTD deletion (INΔNTD), deletion of the N-terminal part of CCD (INΔCCD1), deletion of the C-terminal part of CCD (INΔCCD2), or deletion of the CTD (INΔCTD) were constructed (Fig. 5A). Wild-type IN (WT IN) and IN mutant proteins were expressed efficiently in transfected 293T cells (Fig. 5B, lanes 2 to 6). Expression of A3G-HA proteins in cotransfected 293T cells was also detected (lanes 1 to 6). Immunoprecipitation of WT INmyc and INmyc mutants from cell lysates was achieved using an anti-myc antibody (lanes 8 to 12). IN mutants containing the NTD deletion (lane 9) or deletions in the N-terminal (lane 10) or C-terminal part of the CCD (lane 11) still interacted with A3G-HA, like the wild-type IN (lane 8). A3G-HA was not coprecipitated in the absence of INmyc (lane 7), indicating the specificity of the interactions between A3G-HA and the various IN constructs. However, deletion of the CTD abolished the interaction of the mutant IN with A3G (lane 12). Collectively, these results indicate that A3G and HIV-1 IN form a specific interaction that is dependent on the CTD of IN. A3GC291S was less effective in preventing the formation of proviral DNA than was A3G. We observed that A3GC291S-HA (Fig. 5C, lane 3) also interacted less efficiently with HIV-1 IN-myc than did A3G-HA (Fig. 5C, lane 1).
FIG. 5.
(A) The C terminus of HIV-1 IN and the first linker of A3G are required for the interaction of A3G and IN. HIV-1 IN domains and HIV-1 IN mutants used in this study are shown. (B) HA-tagged A3G expression vector was cotransfected with the INmyc expression vector or a control vector into 293T cells. At 48 h posttransfection, cells were lysed and analyzed by immunoblotting using the anti-HA MAb for the detection of A3G-HA and the anti-myc MAb for the detection of INmyc (lanes 1 to 6). Cell lysates were also immunoprecipitated (IP) by the anti-myc MAb conjugated to protein G-agarose beads. Coprecipitated samples were analyzed by immunoblotting using the anti-HA MAb for the detection of A3G-HA or the anti-myc MAb for the detection of INmyc (lanes 7 to 12). (C) HA-tagged A3G or A3GC291S expression vector was cotransfected with the INmyc expression vector or a control vector into 293T cells. At 48 h posttransfection, cells were lysed and analyzed by immunoblotting using the anti-HA MAb for the detection of A3G-HA and A3GC291S-HA. Cell lysates were also immunoprecipitated by the anti-myc MAb conjugated to protein G-agarose beads. Coprecipitated samples were analyzed by immunoblotting using the anti-HA MAb for the detection of A3G-HA and A3GC291S-HA or anti-myc MAb for the detection of INmyc. (D) The A3G domains and A3G mutants used in this study. (E) A3G or mutant A3G was cotransfected with the myc-tagged IN expression vector or a control vector into 293T cells. At 48 h posttransfection, cells were lysed and analyzed by immunoblotting using the anti-HA and anti-myc MAbs (lanes 1 to 8). Cell lysates were also immunoprecipitated by the anti-myc MAb conjugated to protein G-agarose beads. Coprecipitated samples were analyzed by immunoblotting using the anti-HA MAb or anti-myc MAbs (lanes 9 to 16).
By analogy to APOBEC1, the functional domains of A3G could be separated into an N-terminal domain, first active site, first linker region, pseudoactive site, second active site, second linker region, and C-terminal domain (21). Truncated mutants of human A3G constructs were used to identify regions important for A3G and IN interaction (Fig. 5D). A3G mutant proteins were expressed efficiently in transfected 293T cells (Fig. 5E, lanes 1 to 8). INmyc was also expressed in cotransfected cells (lanes 2, 4, 6, and 8). Immunoprecipitation of INmyc from cell lysates was achieved using an anti-myc antibody (lanes 10, 12, 14, and 16). Deletion of the N-terminal 104 amino acids (lane 10), C-terminal 157 to 384 amino acids (lane 14), or C-terminal 246 to 384 amino acids (lane 16) of A3G did not abolish its ability to interact with IN. However, deletion of the N-terminal 156 amino acids of A3G abolished its interaction with IN (lane 12). This result indicates that amino acids 104 to 156, found in the N-terminal portion of a linker sequence between the two zinc coordination motifs of A3G, are critical for its interaction with IN.
A3C does not interact with NCp7 or IN and does not affect viral reverse transcription or integration.
A3C, a member of the human APOBEC3 family of cytidine deaminases, is less potent against HIV-1 than is A3G (4, 24, 29, 58, 60, 65) and has also been reported to have strong activity against SIV. In order to dissect the antiviral function of A3C, we next quantified the amount of ERT, LRT, 2-LTR, and integrated viral DNA after infection. Unlike A3G, A3C did not significantly affect the reverse transcription of HIV-1 (Fig. 6A). The kinetics and peak levels of ERT and LRT in the presence and absence of A3C were not significantly different. However, the quantity of the 2-LTR viral DNA was fivefold lower in the presence of A3C than in its absence (Fig. 6B). The quantity of the integrated viral DNA was also reduced by approximately fivefold in the presence of A3C (Fig. 6C). Thus, viral integration was apparently not inhibited by A3C. To determine whether NCp7 or INp32 also interacts with A3C proteins, A3C or A3G expression vector was cotransfected with HIV-1ΔVif, and virions were purified from transfected 293T cells. A3C-HA and A3G-HA were immunoprecipitated from comparable amounts of virions (Fig. 6D, lanes 1 to 3), and coprecipitated samples were examined for INp32 or NCp7 (lanes 4 to 6). In contrast to A3G (Fig. 6D, lane 5), A3C-HA (lane 6) did not interact efficiently with HIV-1 IN or NC (lane 6). A3C could induce cytidine deamination in HIV-1 DNA (Fig. 6E).
FIG. 6.
A3C did not interact with NCp7 or IN and did not affect reverse transcription or integration. (A to C) Effects of A3C on the early steps of HIV-1 replication. 293T cells were transfected with the HIV-1ΔVif-based vector and VSV-G plus A3C or control vector. At 3 days posttransfection, HeLa cells were infected with HIV-1ΔVif that had been pretreated with DNase I. Total DNA was isolated at the indicated times following infection, and ERT, LRT, and 2-LTR circle DNAs were quantified. Virus input was normalized according to the level of p24. The highest value at peak production of HIV-1 from 293T cells cotransfected with pcDNA3.1 was set to 100%. (A) Quantification of ERT products and LRT products. (B) Quantification of 2-LTR circles. (C) Quantification of integration at 24 h after infection. (D) A3C did not interact with HIV-1 IN or NCp7 in released virions. HIV-1ΔVif was cotransfected with the pcDNA3.1, A3G-HA, or A3C-HA expression vector into 293T cells. At 72 h posttransfection, viruses were collected and purified from culture supernatants. Viral lysates were analyzed by immunoblotting using pooled HIV-1-positive human sera for the detection of viral proteins or an anti-HA MAb for the detection of virion-packaged A3G-HA or A3C-HA (lanes 1 to 3). Viral lysates were also immunoprecipitated (IP) with the anti-HA MAb conjugated to agarose beads, and immunoprecipitated samples were also analyzed by immunoblotting using an anti-HIV-1 IN antibody, anti-HIV-1 p7 antibody, and anti-HA MAb (lanes 4 to 6). (E) HIV-1 sequences in the region of the Nef-U5 fragment from virus-infected cultures. G-to-A mutations are shown. nt, nucleotide.
Antiviral A3F interacts with IN and inhibits the formation of proviral DNA.
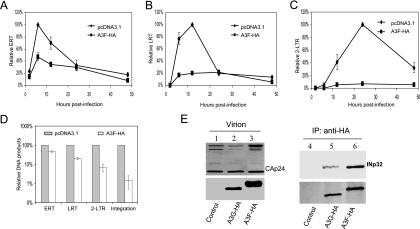
A3F is also effective against HIV-1 (4, 24, 29, 58, 60, 65). In order to dissect the antiviral function of A3F, we next quantified the amount of ERT, LRT, 2-LTR, and integrated viral DNA after infection. Like A3G, A3F also reduced the reverse transcription of HIV-1 (Fig. 7A and B) and the formation of proviral DNA (Fig. 7D). Interaction between INp32 and A3F-HA in HIV-1ΔVif virions was also detected (Fig. 7E, lane 6).
FIG. 7.
Effects of A3F on the early stages of HIV-1 replication during a single-round infection of HeLa cells. (A to D) HIV-1ΔVif was cotransfected with the pcDNA3.1 or A3F-HA expression vector and VSV-G into 293T cells. At 3 days posttransfection, HeLa cells were infected with HIV-1ΔVif that had been pretreated with DNase I. Total DNA was isolated from the infected cells at the indicated times following infection, and early RT, late RT, and 2-LTR circles were quantified. Virus input was normalized according to the level of p24. The highest value at peak productivity of HIV-1 from 293T cells cotransfected with pcDNA3.1 was set to 100%. (A) Quantification of early RT products. (B) Quantification of late RT products. (C) Quantification of 2-LTR circles. (D) Comparison of ERT, LRT, 2-LTR, and integration (Alu-based qRT-PCR) in the absence or presence of A3F-HA at the peak points. (E) HIV-1ΔVif was cotransfected with the pcDNA3.1, A3G-HA, or A3F-HA expression vector into 293T cells. At 48 h posttransfection, viruses were collected and purified from culture supernatants. Viral lysates were analyzed by immunoblotting using pooled HIV-1-positive human sera for the detection of viral proteins (lanes 1 to 3, upper panel) or an anti-HA MAb for the detection of virion-packaged A3G-HA or A3F-HA (lanes 1 to 3, lower panel). Viral lysates were also immunoprecipitated (IP) with the anti-HA MAb conjugated to agarose beads, and coprecipitated samples were then analyzed by immunoblotting using the anti-INp32 antibody (lanes 4 to 6, upper panel) or the anti-HA MAb to detect immunoprecipitated A3G-HA and A3F-HA (lanes 4 to 6, lower panel).
DISCUSSION
A major finding of the current study is the antiviral effect of cytidine deaminases A3G and A3F on the formation of HIV-1 proviral DNA. At levels similar to those of endogenous A3G in H9 cells, the most potent antiviral activity of A3G occurred at the step of viral DNA integration. A3G, A3F, and, to a lesser extent, A3GC291S were able to inhibit the formation of HIV-1 proviral DNA, whereas another human cytidine deaminase, A3C, showed no such antiviral activity. A3G and A3F, but not A3C, interacted with IN and NC in released HIV-1 virions. Association of A3G with a component(s) of the preintegration complex (PIC) such as IN may interfere with the structural integrity of PIC and consequently inhibit PIC intracellular transport or integration of the viral DNA. The interaction of A3G with IN maps to the CTD of IN. While the catalytic site responsible for viral DNA processing and host DNA ligation lies in the central catalytic domain (2, 10, 27), evidence also suggests that the CTD of IN contributes to the integration of HIV-1 DNA (10, 28). Therefore, A3G could also interfere with IN function and viral DNA integration through binding to the CTD of IN.
Since A3G was more effective than A3GC291S in blocking the formation of proviral DNA, the ability to generate uracil in viral DNA may influence the potency of A3G in terms of its ability to inhibit viral integration. However, sequence analysis of 2-LTR DNA junctions in the presence of A3G did not reveal extensive G-to-A hypermutations near the viral DNA attachment sites or deletions indicating degradation of the viral DNA ends (data not shown). Alternatively, it is possible that the C-terminal catalytic domain of A3G is involved in binding to viral or cellular factors that inhibit viral integration and that A3GC291S has a weaker effect on viral integration because of its reduced ability to interact with these factors. Consistent with this idea, we observed that the ability of A3GC291S to interact with HIV-1 IN was lower than that of A3G. Binding of A3G to single-stranded DNA has been demonstrated (20, 60). Whether A3G can bind to viral DNA and block integration by inhibiting the recognition of viral DNA ends by IN is, however, an open question.
Expression of A3G and A3GC291S also reduced the accumulation of viral DNA products, presumably through inhibition of reverse transcription (DNA synthesis) or enhancement of viral DNA degradation. Consistent with previous observations (36, 37), the reduction in viral DNA levels could not fully account for the antiviral activity of A3G (Fig. 3E). Since the disappearance of viral DNA was similar in the presence and absence of A3G and the catalytic mutant A3GC291S was still able to decrease the accumulation of viral DNA, it is less likely that host DNA repair enzyme-mediated degradation of viral DNA is the only cause of the reduced accumulation of viral DNAs. The degradation-independent reduction in HIV-1 reverse transcription by A3G has also been proposed recently by other groups (3, 17).
A noticeable difference between deamination-competent A3G and deamination-defective A3GC291S was that A3G also reduced the accumulation of 2-LTR circle DNA. A3C, which has only weak antiviral activity, mainly reduced the accumulation of 2-LTR circle DNA. A3G and A3C, but not A3GC291S, was able to induce cytidine deamination of viral DNA. It is possible that uracil-containing DNAs were not transported efficiently to the nucleus to form 2-LTR circle DNA. Alternatively, uracil-containing DNAs, especially those near the ends of viral DNA, might not have been recognized efficiently by the DNA ligase in the nucleus and thus inhibited the formation of 2-LTR circle DNA. Furthermore, it could be argued that uracil-containing DNAs are degraded more rapidly in the nucleus, and therefore the reduction in 2-LTR circle DNA (only in the nucleus) would be greater than that of the LRT products (present in both the cytoplasm and nucleus). Whether the host's DNA repair machinery is involved in the degradation of HIV-1 uracil-containing DNAs is an open question. The reduction in LRT products (Fig. 3C) and 2-LTR circle DNA (Fig. 3D) in the presence of A3G was observed at both early and later time points after infection, and there was no evidence that these DNAs disappeared more rapidly. Expression of UGI, an inhibitor of UNG2 (57), in virus-producing cells or target cells also did not reduce the antiviral activity of A3G (data not shown). Thus, further studies will be required to address the possibility that DNA degradation is responsible for the reduction in reverse transcription products or the 2-LTR circle DNA.
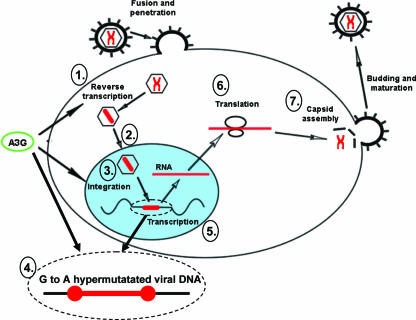
In summary, we have shown that A3G and A3F can mediate HIV-1 suppression through multiple mechanisms, including interference with reverse transcription and integration during HIV-1 replication (Fig. 8). Through its interaction with NCp7 and INp34, multiple steps of the HIV-1 replication cycle could be inhibited by both cytidine deamination-dependent and -independent mechanisms. The most potent antiviral activity of A3G occurred at the step of proviral DNA formation after reverse transcription and nuclear localization of viral DNA. A further understanding of these A3G-mediated antiviral mechanisms may offer opportunities for the design of more effective HIV-1 inhibitors.
FIG. 8.
Model for the multiple effects of A3G activity on HIV-1 replication. A3G can affect HIV-1 replication steps, including accumulation of reverse transcription products (1), proviral DNA formation (3), and proviral DNA integrity by G-to-A hypermutation (4). Note that these affected processes occur early in the HIV-1 life cycle, whereas transcription (5), translation (6), and virus assembly (7) were not affected by A3G.
Acknowledgments
We thank P. T. Sarkis, E. Ehrlich, and R. Markham for advice and technical assistance, M. Malim, R. Gorelick, S. Cen, and L. Kleiman for critical reagents, and D. McClellan for editorial assistance. MAGI-CCR5 cells, anti-HIV-1 integrase, and anti-human A3G antibodies were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by a grant from the NIH (AI062644) and funding from the National Science Foundation of China (NSFC-30425012) as well as Cheung Kong Scholars Program Foundation of the Chinese Ministry of Education to X.-F. Yu.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, N. J. 2004. HIV-1 integrase: a target for new AIDS chemotherapeutics. Curr. Top. Med. Chem. 4:979-990. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, S. O'Shea, K. J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 8.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480-485. [DOI] [PubMed] [Google Scholar]
- 10.Chiu, T. K., and D. R. Davies. 2004. Structure and function of HIV-1 integrase. Curr. Top. Med. Chem. 4:965-977. [DOI] [PubMed] [Google Scholar]
- 11.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435:108-114. [DOI] [PubMed] [Google Scholar]
- 12.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 13.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281-288. [DOI] [PubMed] [Google Scholar]
- 14.Douaisi, M., S. Dussart, M. Courcoul, G. Bessou, R. Vigne, and E. Decroly. 2004. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem. Biophys. Res. Commun. 321:566-573. [DOI] [PubMed] [Google Scholar]
- 15.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 17.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 19.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10:1247-1253. [DOI] [PubMed] [Google Scholar]
- 20.Iwatani, Y., H. Takeuchi, K. Strebel, and J. G. Levin. 2006. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 80:5992-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, M., A. Takaori-Kondo, Y. Miyauchi, K. Iwai, and T. Uchiyama. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 280:18573-18578. [DOI] [PubMed] [Google Scholar]
- 23.Kong, W., C. Tian, B. Liu, and X. F. Yu. 2002. Stable expression of primary human immunodeficiency virus type 1 structural gene products by use of a noncytopathic Sindbis virus vector. J. Virol. 76:11434-11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 26.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 27.Lewinski, M. K., and F. D. Bushman. 2005. Retroviral DNA integration-mechanism and consequences. Adv. Genet. 55:147-181. [DOI] [PubMed] [Google Scholar]
- 28.Li, M., and R. Craigie. 2005. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J. Biol. Chem. 280:29334-29339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 30.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, B., X. Yu, K. Luo, Y. Yu, and X. F. Yu. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lochelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y. B. Kim, U. Truyen, U. Rosler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Munk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo, K., Z. Xiao, E. Ehrlich, Y. Yu, B. Liu, S. Zheng, and X. F. Yu. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. USA 102:11444-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahieux, R., R. Suspene, F. Delebecque, M. Henry, O. Schwartz, S. Wain-Hobson, and J. P. Vartanian. 2005. Extensive editing of a small fraction of human T-cell leukemia virus type 1 genomes by four APOBEC3 cytidine deaminases. J. Gen. Virol. 86:2489-2494. [DOI] [PubMed] [Google Scholar]
- 36.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 37.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 38.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 39.Mehle, A., J. Goncalves, M. Santa-Marta, M. McPike, and D. Gabuzda. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18:2861-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 41.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 42.Noguchi, C., H. Ishino, M. Tsuge, Y. Fujimoto, M. Imamura, S. Takahashi, and K. Chayama. 2005. G to A hypermutation of hepatitis B virus. Hepatology 41:626-633. [DOI] [PubMed] [Google Scholar]
- 43.Rose, P. P., and B. T. Korber. 2000. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics 16:400-401. [DOI] [PubMed] [Google Scholar]
- 44.Rosler, C., J. Kock, M. H. Malim, H. E. Blum, and F. von Weizsacker. 2004. Comment on “Inhibition of hepatitis B virus replication by APOBEC3G.” Science 305:1403; author reply 1403. [DOI] [PubMed] [Google Scholar]
- 45.Sasada, A., A. Takaori-Kondo, K. Shirakawa, M. Kobayashi, A. Abudu, M. Hishizawa, K. Imada, Y. Tanaka, and T. Uchiyama. 2005. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 47.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 49.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 50.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 51.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 52.Stevenson, M. 2002. Molecular biology of lentivirus-mediated gene transfer. Curr. Top. Microbiol. Immunol. 261:1-30. [DOI] [PubMed] [Google Scholar]
- 53.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 54.Suspene, R., P. Sommer, M. Henry, S. Ferris, D. Guetard, S. Pochet, A. Chester, N. Navaratnam, S. Wain-Hobson, and J. P. Vartanian. 2004. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 32:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 56.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Response to comment on “Inhibition of hepatitis B virus replication by APOBEC3G.” Science 305:1403b. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Z., and D. W. Mosbaugh. 1989. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J. Biol. Chem. 264:1163-1171. [PubMed] [Google Scholar]
- 58.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 60.Yu, Q., R. Konig, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 61.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 62.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]