Abstract
The search for hepatitis C virus polymerase inhibitors has resulted in the identification of several nonnucleoside binding pockets. The shape and nature of these binding sites differ across and even within diverse hepatitis C virus genotypes. These differences confront antiviral drug discovery with the challenge of finding compounds that are capable of inhibition in variable binding pockets. To address this, we have established a hepatitis C virus mutant and genotypic recombinant polymerase panel as a means of guiding medicinal chemistry through the elucidation of the site of action of novel inhibitors and profiling against genotypes. Using a genotype 1b backbone, we demonstrate that the recombinant P495L, M423T, M414T, and S282T mutant enzymes can be used to identify the binding site of an acyl pyrrolidine analog. We assess the inhibitory activity of this analog and other nonnucleoside inhibitors with our panel of enzyme isolates generated from clinical sera representing genotypes 1a, 1b, 2a, 2b, 3a, 4a, 5a, and 6a.
Hepatitis C is estimated to affect 3% of the global population. In a number of individuals, it can lead to liver fibrosis, cirrhosis, and death. Although virus can be cleared by a combination of pegylated interferon and ribavirin, the treatment is successful in only around 50% of treated patients and has considerable liabilities. These weaknesses highlight the need for new drugs to treat hepatitis C virus (HCV) in patients who have failed current therapy, as well as in untreated patients (12, 56).
HCV is an enveloped virus with an RNA genome of ∼9.6 kb. Its single-stranded RNA has a positive polarity and encodes a polyprotein of ∼3,300 amino acids comprising 4 structural proteins (Core, E1, E2, and p7) and 6 nonstructural proteins (NS2, -3, -4A, -4B, -5A, and -5B) (43). These proteins, as well as the viral translation process using the internal ribosomal entry site and a range of host factors, are candidate targets for therapeutic intervention (3, 46). The remarkable clinical success of human immunodeficiency virus reverse transcriptase and protease inhibitors, as well as the availability of several crystal structures, has motivated HCV drug discovery efforts to focus mainly on the development of protease and polymerase inhibitors. HCV NS5B is an RNA-dependent RNA polymerase that is responsible for the replication of the viral genome, which is thought to occur through a primer-independent de novo mechanism (6, 31). Due to the lack of proofreading capacity, this replication process is subject to a high error rate (36). As a result, the virus has evolved into multiple variant strains, classified into six different genotypes (1 to 6) and several subtypes (a, b, c, etc.) (45). To add to this complexity, HCV-infected individuals also harbor different variants or quasispecies of the virus, together representing a pool of genomes on which selective pressure can act (16). It has been speculated that drug resistance will rapidly emerge upon administration of specific HCV antivirals and that together with viral genotype, these issues will be important in the pursuit of effective therapies.
As for other polymerases, HCV has adopted a generic topology for NS5B, i.e., a right-hand motif consisting of a thumb domain and a fingers domain, which encircle the active site located within the palm domain (26). NS5B inhibitors can be classified into nucleoside and nonnucleoside inhibitors (NIs and NNIs, respectively) (9, 32, 44, 50). NIs resemble nucleosides, which act by competing with the natural ribonucleoside triphosphate substrates of NS5B at its catalytic center. NNIs are chemically diverse and inhibit the initiation and/or elongation step by binding near the active site or discrete allosteric sites. To date, at least three distinct inhibitor binding sites have been reported, NNI-1, -2, and -3 (see Fig. 1) (9, 10). The NNI-1 site is located on the surface of the thumb domain adjacent to the allosteric GTP site (4, 13). Ligands identified against this site include both benzimidazole (1, 51) and indole derivatives (13, 20). The NNI-2 site is located in the thumb domain, next to NNI-1 (2, 29, 55). Chemotypes of NNI-2 binders include the thiophene (2, 7), phenylalanine (8), dihydropyranone (29), and pyranoindole analogs (17). The NNI-3 site is located adjacent to the active site. Reported NNI-3 ligands include benzothiadiazine (11, 47), proline sulfonamide (18), benzylidene (24, 42), and acrylic acid (40, 41) derivatives. In drug discovery, knowledge of the inhibitor site of action is crucial to guiding medicinal chemistry efforts. Structural activity relationships are further complicated by the variation observed for each of the NNI binding sites between genotype and subtypes. These issues can be addressed using X-ray crystallography, as demonstrated by others (2, 13, 18, 24, 29, 40, 41, 42, 47, 55). However, this is a considerable undertaking and requires both time and abundant purified enzyme.
FIG. 1.
Structural representation of the HCV NS5B polymerase. Palm, fingers and thumb subdomain of NS5B are color coded red, green, and blue, respectively. Magnesium atoms are shown as purple spheres. The chemical structures of the selected reference compounds benzimidazole 1, thiophene 2, benzothiadiazine 3, and the inhibitor to which the binding site was unknown, acyl pyrrolidine 4, are represented next to the polymerase structure. An arrow indicates the NNI binding site of each reference compound, 1, 2, and 3. The areas color coded magenta, yellow, or orange represent the binding site of our reference compound 1, 2, or 3 based on reported X-ray crystallography studies (PDB 2BRK, 1OS5, and 1YVF, respectively). Mutants observed in resistance selection experiments are mentioned in brackets next to the binding site. For a complete mutant panel, we also included the S282T mutant of the active site, located near the catalytic aspartate residues.
Here we have developed an NS5B enzyme panel comprising both site-directed mutants and representatives from different genotypes to elucidate the site of action of novel inhibitors and to profile their inhibitory activities across genotypes. We describe the use of the panel to provide insights into the rational drug design of inhibitors that can be accommodated into variable binding sites.
MATERIALS AND METHODS
Materials and compounds.
Custom-made oligonucleotides and 5′-biotinylated oligo(rG13) were ordered from Sigma-Proligo (France). The poly(rC) template was obtained from GE HealthCare (Uppsala, Sweden). Restriction endonucleases were purchased from New England BioLabs (Ipswich, MA). Plasmid DNA isolation was performed using either the Miniprep or Midiprep plasmid purification kit from QIAGEN (Hilden, Germany). PCR products were purified using the QIAGEN (Hilden, Germany) PCR purification kit. Ligations were performed using the Rapid DNA ligation kit from Roche (Basel, Switzerland). Proteins were purified on an ÄKTAxpress purification module (GE HealthCare, Uppsala, Sweden) using chromatographic equipment from the same manufacturer.
The following reference compounds were synthesized as described: compound 1, a benzimidazole scaffold (N-{[1-cyclohexyl-2-(3-furyl)-1H-benzimidazol-5-yl]carbonyl}-l-tryptophan) (1), compound 2, a thiophene scaffold (3-[isopropyl(trans-4-methylcyclohexylcarbonyl)amino]-5-phenylthiophene-2-carboxylic acid) (7), compound 3, a benzothiadiazine scaffold {3-(1,1-dioxo-1,4-dihydrobenzo[1,2,4]thiadiazin-3-yl)-4-hydroxy-1-(3-methylbutyl)-1H-quinolin-2-one} (11, 35), compound 4, an acyl pyrrolidine scaffold [rel-(2S,4S,5R)-2-isobutyl-1-(4-tert- butylbenzoyl)4-cyano-5-(1,3-thiazol-2-yl)pyrrolidine-2-carboxylic acid] (G. Burton, H. S. Goodland, D. Haigh, T. J. Kiesow, T. W. Ku, and M. J. Slater, 1-Carbonyl-4-cyano-pyrrolidine-2-carboxylic acid derivatives as hepatitis C inhibitors, international patent application WO 2004009543 A2, 2004).
PCR amplification of NS5B genes from clinical isolates.
Serum samples were genotyped using the Line Probe Assay HCV II genotyping kit (Versant HCV, Bayer Diagnostic, Berkeley, CA). For this, total RNA was first extracted using the QIAamp viral RNA kit from QIAGEN (Hilden, Germany). Serum samples of genotypes 4 to 6 were purchased from Teragenix (Fort Lauderdale, FL). Samples were classified as either genotype 1a, 1b, 2a, 2b, 3a, 4a, 5a, or 6a. Isolated RNA was subsequently transcribed into cDNA with ImProm-II reverse transcriptase (Promega, Madison, WI) using a subtype-specific primer located in the 3′ untranslated region of the viral RNA (Table 1). The resulting cDNA was amplified by means of a nested PCR protocol, using the Expand Long Template PCR system (Roche, Basel, Switzerland); forward primers were designed based on the coding sequence for the structural protein E2 and the reverse primer within the 3′ untranslated region.
TABLE 1.
Primers used in this study
| Identifier | Sequencea |
|---|---|
| Subtype-specific primers used in reverse transcription | |
| HCV1a-RT | 5′-AGG CCG GAG TGT TTA CCC CAA C-3′ |
| HCV1b-RT | 5′-GGC CTG GAG TGG TTA GCT CCC C-3′ |
| HCV2a-RT | 5′-GTT AGC TAT GGA GTG TAG CTA-3′ |
| HCV2b-RT | 5′-ATG GAG TGT AGC TAG GGT TTG CC-3′ |
| HCV3a-RT | 5′-GAA ATG GAG TGT TAT CCT ACC-3′ |
| HCV4a-RT | 5′-CCT AAG GTC GGA GTG TTA AG-3′ |
| HCV5a-RT | 5′-GGA GTG TTT AGC TCC CAG C-3′ |
| HCV6a-RT | 5′-GAG TGT TGC TAA GGC CGC TCG-3′ |
| Primers used for NS5B cloning | |
| FP1b_J4 | 5′-TAG GGC GCT AGC TCA ATG TCC TAT ACG TGG ACA GGC GCC-3′ |
| RP1b_J4 | 5′-TAG GGC CTC GAG GCG GGG TCG GGC ACG AGA CAG GCT GTG-3′ |
| FP1a | 5′-TAG GGC GCT AGC TCA ATG TCT TAT TCC TGG-3′ |
| RP1a | 5′-TAG GGC CTC GAG GCG GGG CCG GGC ATG-3′ |
| FP1b | 5′-TAG GGC GCT AGC TCA ATG TCC TAC ACA TGG-3′ |
| RP1b | 5′-TAG GGC CTC GAG GCG GGG TCG GGC ACG-3′ |
| FP2a | 5′-TAG GGC GCT AGC TCC ATG TCA TAC TCC TGG-3′ |
| RP2b | 5′-TAG GGC CTC GAG GCG GGG TCG GGC ATG-3′ |
| FP3a | 5′-TAG GGC GCT AGC TCT ATG TCG TAC TCT TGG-3′ |
| RP3a | 5′-TAG GGC CTC GAG GCG GGT TCG GGC ACG-3′ |
| FP4a | 5′-TAG GGC GCT AGC TCG ATG TCA TAC TCG TGG AC-3′ |
| RP4a | 5′-TAG GGC CTC GAG GCG GGG TCG GGC ATG-3′ |
| FP5a | 5′-TAG GGC CAT ATG TCC ATG TCA TAC ACC TGG ACT-3′ |
| RP5a | 5′-TAG GGC GCG GCC GCG CGG GGT CGG GCA CG-3′ |
| FP6a | 5′-TAG GGC CAT ATG TCC ATG TCC TAT TCC TGG ACG-3′ |
| RP6a | 5′-TAG GGC GCG GCC GCA CGG GGT CGG GCC TG-3′ |
NheI, XhoI, NdeI, and NotI sites of primers used for NS5B cloning are shown in bold.
Cloning, expression, and purification of NS5B.
The coding sequence for the NS5B (genotype 1b consensus strain J4) lacking 21 C-terminal residues was amplified from plasmid pCV-J4L6S (GenBank accession no. AF054247) using Deep Vent DNA polymerase (New England BioLabs, Ipswich, MA) and a set of primers containing either an NheI site or an XhoI site (Table 1). The PCR product was purified and digested with NheI/XhoI prior to cloning into the NheI/XhoI-digested pET21b plasmid (Novagen, Madison, WI) to generate the expression plasmid for NS5BΔ21.
Plasmids encoding mutant NS5BΔ21 proteins (P495L, M423T, M414T, and S282T) in which Pro495, Met423, Met414, and Ser282 are individually replaced with leucine or threonine were generated with the QuickChange site-directed mutagenesis kit from Stratagene (La Jolla, CA), using the 1b J4 NS5BΔ21 expression plasmid and specific DNA oligonucleotides. All resulting expression vectors were verified by DNA sequencing.
The NS5B sequences from the clinical isolates were amplified from their respective cDNA templates (described above) using Deep Vent DNA polymerase (New England BioLabs, Ipswich, MA) and oligonucleotides listed in Table 1. The resulting PCR products were cloned into pET21b as described above to generate His-tagged expression constructs, each harboring the C-terminal 21-residue deletion as in the plasmid NS5BΔ21. Among the cloned NS5B sequences, we selected two 1a and two 1b representatives (1a_2, 1a_6, 1b_9, and 1b_10) and one 2a (2a_11), one 2b (2b_12), two 3a (3a_13 and 3a_14), one 4a (4a_26), one 5a (5a_29), and one 6a (6a_28) representative (see Fig. 3).
FIG. 3.
Comparative amino acid sequence analysis of NS5B and unrooted phylogenetic tree representation of NS5BΔ21 from 11 clinical isolates and genotype 1b consensus strain J4. Percent identities and similarities are determined according to the BLOSUM62 algorithm. Sequence length is without the C-terminal His tag and initiating residues. Masses are calculated with exclusion of the C-terminal His tag and initiating residues. Phylogenetic tree based on the multiple sequence alignment of NS5BΔ21 from 1b J4 and 11 clinical isolates. The tree was visualized with TREEVIEW. Numbers represent bootstrap values.
The NS5B expression constructs were transformed into Escherichia coli BL21(DE3) (Novagen, Madison, WI). Five milliliters of LB medium supplemented with ampicillin (50 μg/ml) was inoculated with one colony. When the preculture reached an optical density of 0.6 measured at 600 nm, it was transferred to fresh LB medium supplemented with ampicillin at a ratio of 1:200. Cells were grown at 37°C to an optical density at 600 nm of 0.6 and then shifted to a growth temperature of 20°C following induction with ispopropyl-1-thio-β-d-galactopyranoside and MgCl2 at final concentrations of 0.4 mM and 10 μM, respectively. After 10 h of induction, cells were harvested by centrifugation and resuspended in 20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 10% glycerol, 0.1% NP-40, 4 mM MgCl2, and 5 mM dithiothreitol (DTT) supplemented with EDTA-free complete protease inhibitor (Roche, Basel, Switzerland). Cell suspensions were disrupted by sonication and incubated with 10 to 15 mg/liter of DNase I (Roche, Basel, Switzerland) for 30 min. Cell debris was removed through ultracentrifugation at 30,000 × g for 1 h, and clarified cell lysate was flash frozen and stored at −80°C prior to purification.
Clarified cell lysate was thawed and loaded onto a 5-ml prepacked HisTrap FF column equilibrated with 25 mM HEPES, pH 7.5, 500 mM NaCl, 10% glycerol, and 5 mM DTT. Proteins were eluted with 500 mM imidazole at a flow rate of 1 ml/min. Fractions of interest were applied to a prepacked 26/10 HiPrep desalting column equilibrated with 25 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, and 5 mM DTT. The buffer-exchanged NS5B peak was then applied to a 20 ml Poly-U Sepharose column. Protein was eluted with an increasing salt gradient and fractions collected. Protein purity was assessed on Nu-PAGE precast gels (Invitrogen, Carlsbad, CA). Purified NS5B samples were concentrated using Centri-Prep concentrators (Millipore, Billerica, MA), and protein concentrations were determined by using the Bradford assay (Pierce, Rockford, IL).
RNA-dependent RNA polymerase assay.
Fifty-percent inhibitory concentrations (IC50s) were essentially determined as described previously (15) using a primer-dependent transcription assay. Briefly, 100 nM of purified NS5B enzyme was incubated with 300 nM 5′-biotinylated oligo(rG13)/poly(rC) primer template, 600 nM of GTP, and 0.1 μCi of [3H]GTP in 25 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, 17 mM NaCl, and 3 mM of DTT. The 30-μl reaction mixture was incubated at room temperature for 2 h before the reaction was stopped by addition of 30 μl of streptavidin-coated SPA beads (GE Healthcare, Uppsala, Sweden) in 0.5 M EDTA. Next, the 384-well plates were counted by scintillation proximity on a Packard TopCount reader.
Similar conditions were employed for the determination of the kinetic parameters of each NS5B enzyme, apart from the following modifications: while keeping the primer/template concentration constant, we varied the amount of [3H]GTP between 0 and 4.8 μM. For the primer/template titration, we used a [3H]GTP concentration of 4.8 μM while varying the primer/template concentration between 0 and 300 nM. All experiments were performed at least twice in quadruplicate, and kinetic parameters were calculated using nonlinear regression in GraphPad Prism 4.0 (GraphPad Software, San Diego, CA).
Phylogenetic analysis.
Amino acid sequences were aligned using ClustalX (49). The alignment was subjected to the bootstrap procedure of the PHYLIP package 3.61 (14). After 100 bootstrap cycles, the data set was used to calculate the distances using the PROTDIST method with the Dayhoff PAM001 matrix. This output was further subjected to the least-squares method using FITCH to search for the best trees. The produced series of trees was analyzed by the CONSENSE program to reveal the most probable tree, which was used as a “guide tree.” The tree was visualized with TREEVIEW (38).
Nucleotide sequence accession numbers.
The NS5B sequences derived from the clinical isolates and described in this work have been deposited in GenBank and assigned accession numbers EF523591 (1a_2), EF523592 (1a_6), EF523593 (1b_9), EF523594 (1b_10), EF523595 (2a_11), EF523596 (2b_12), EF523597 (3a_13), EF523598 (3a_14), EF523599 (4a_26), EF523600 (5a_29), and EF523601 (6a_28).
RESULTS
Use of a mutant panel to identify a binding site.
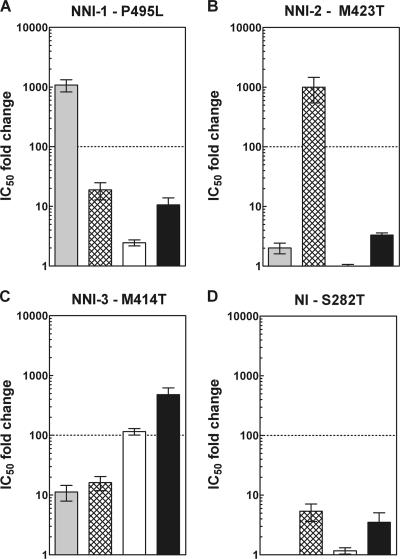
From the several classes of NNI inhibitors that have been reported, we selected and synthesized benzimidazole 1 (1), thiophene 2 (7), and benzothiadiazine 3 (11, 35) as reference compounds for each NNI site (Fig. 1). We also synthesized acyl pyrrolidine 4, reported as an NS5B inhibitor with an uncharacterized binding site (5; Burton et al., international patent application WO 2004009543 A2, 2004). Resistance selection experiments in HCV replicon cells revealed that the P495L mutation in NS5B was sufficient to confer resistance to benzimidazoles (23, 50, 51), the M423T mutation conferred resistance to thiophenes (10, 25, 50), and the M414T mutation conferred resistance to benzothiadiazines (34, 35, 50, 52) (Fig. 1). The observation that each replicon NS5B mutant was resistant only to its cognate inhibitor suggested that recombinant mutant enzymes could be used as a panel to elucidate the site of action of novel inhibitors (30, 50). To address this, we generated overexpression constructs of these mutants from the parental NS5BΔ21 (J4, genotype 1b) plasmid. In addition, we included the S282T mutant of NS5B, which resists inhibition by 2′-C-methyl nucleosides (33, 50), in order to discriminate between an NI and an NNI. Each mutant polymerase was assessed for its efficiency using GTP and oligo(rG13)/poly(rC) as a primer/template. As expected, Km values for the nucleotide and RNA substrate were within the expected micromolar and nanomolar ranges, respectively, (Table 2). Specific activities of the mutant enzymes were found to be generally comparable to that of the wild-type 1b J4 polymerase. The M414T and S282T mutants have a slightly lower affinity for GTP, possibly caused by their mutation next to the active site. The slight decrease in specific activity for the P495L mutant compared to the wild-type enzyme and the observed increase for the M414T mutant are consistent with previous reports (51, 52).
TABLE 2.
Kinetic parameters for the NS5BΔ21 (1b J4) enzyme, mutants, and clinical isolates
| NS5B protein | Km for GTP (μM) | Km for RNAa (nM) | Sp act (pmol/ min/mg) |
|---|---|---|---|
| NS5BΔ21 (1b J4) | 0.38 ± 0.02 | 39.99 ± 2.70 | 978.60 ± 58.49 |
| NS5BΔ21 (1b J4) P495L | 0.41 ± 0.03 | 39.41 ± 1.20 | 686.28 ± 12.80 |
| NS5BΔ21 (1b J4) M423T | 0.37 ± 0.03 | 45.51 ± 1.40 | 940.94 ± 50.51 |
| NS5BΔ21 (1b J4) M414T | 0.57 ± 0.04 | 68.26 ± 4.50 | 5317.36 ± 191.12 |
| NS5BΔ21 (1b J4) S282T | 0.93 ± 0.07 | 46.01 ± 2.92 | 615.86 ± 32.20 |
| NS5BΔ21 (1a_2) | 0.36 ± 0.04 | 84.22 ± 5.84 | 173.27 ± 5.56 |
| NS5BΔ21 (1a_6) | 0.24 ± 0.05 | 38.55 ± 0.43 | 65.11 ± 10.87 |
| NS5BΔ21 (1b_9) | 0.26 ± 0.02 | 36.37 ± 6.76 | 540.28 ± 34.40 |
| NS5BΔ21 (1b_10) | 0.60 ± 0.05 | 25.93 ± 3.34 | 766.22 ± 73.07 |
| NS5BΔ21 (2a_11) | 0.38 ± 0.05 | 29.42 ± 1.30 | 330.16 ± 13.40 |
| NS5BΔ21 (2b_12) | 0.43 ± 0.08 | 33.38 ± 6.19 | 137.70 ± 2.10 |
| NS5BΔ21 (3a_13) | 0.39 ± 0.03 | 22.02 ± 1.82 | 173.04 ± 12.01 |
| NS5BΔ21 (3a_14) | 0.64 ± 0.12 | 28.33 ± 7.66 | 34.82 ± 1.52 |
| NS5BΔ21 (4a_26) | 0.48 ± 0.08 | 84.61 ± 11.43 | 341.29 ± 26.18 |
| NS5BΔ21 (5a_29) | 0.66 ± 0.19 | 45.51 ± 1.24 | 52.03 ± 5.66 |
| NS5BΔ21 (6a_28) | 0.24 ± 0.09 | 55.18 ± 6.29 | 85.14 ± 11.23 |
RNA template is oligo(rG13)/poly(rC).
Using the same primer-dependent extension assay, we determined the IC50s of selected inhibitors for each mutant enzyme, which were then compared with the value measured for the wild-type 1b J4 enzyme and reported as n-fold changes in the IC50 (Fig. 2). As expected, compounds 1 to 3 inhibited each enzyme except for their cognate mutated protein (Fig. 2). For example, the IC50 for benzimidazole 1 was more than 1,000-fold higher for the P495L mutant than for the wild type (Fig. 2). These results were consistent with the resistance selection data obtained with HCV replicon cells with these classes of reference compounds (25, 35, 50, 51, 52). Next, we used our mutant panel to identify the acyl pyrrolidine 4 binding site. Figure 2 shows that pyrrolidine inhibition was most affected by the M414T mutant, suggesting that the compound binds within the NNI-3 binding site.
FIG. 2.
Inhibitory activities of compounds 1 to 4 for the NS5B mutant panel. (A to D) In vitro activity of the NS5BΔ21 P495L, M423T, M414T, or S282T mutant with compounds 1 to 4. IC50 n-fold changes are calculated with respect to the obtained IC50 of NS5BΔ21 1b J4 for each class of compound. Light-gray bars, benzimidazole 1; Cross-hatched bars, thiophene 2; white bars, benzothiadiazine 3; black bars, N-acyl pyrrolidine 4. Compounds with an IC50 n-fold change greater than 100 were considered to be inactive (dotted line).
HCV RNA-dependent RNA polymerases from clinical isolates show different NNI susceptibilities.
Given the influence of HCV viral genotype on the clinical response to current therapy, it is important to test drug candidates against a spectrum of circulating genotypes. Reverse genetic analysis of replicons (30) is one means of confirming if the inhibitors identified by enzymatic screening will also be active against clinical isolates of HCV. However, despite the functional relevance of the replicon strategy, cell culture efficacy studies to date have a limited genotypic coverage (1 and 2a). A second approach relies on the use of stable cell lines expressing NS5B only (22), but the process becomes time-consuming when more than one genotypic variant is considered. In addition, both strategies cannot be used for compounds that do not penetrate the cells. Therefore, we decided to generate recombinant proteins from clinical sera representing different genotypes as a complementary tool. The NS5B genes from a panel of genotyped sera were cloned as expression vectors encoding C-terminally truncated (Δ21) polymerase enzymes for E. coli expression. Phylogenetic analysis of the amino acid sequences of our enzyme isolates among subtypes and across genotypes was consistent with the sequence information available in the public database (euHCVdb [http://euhcvdb.ibcp.fr]) (Fig. 3). As expected, we observed divergence within a subtype of less than 5% and within a genotype of around 15%, whereas divergence across genotypes was typically 25 to 30% at the amino acid level.
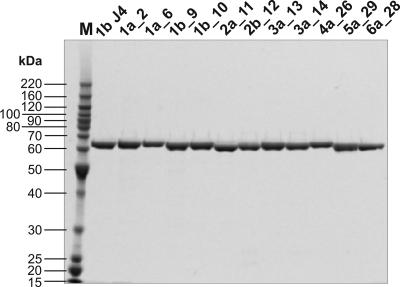
Figure 4 shows sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the purified recombinant NS5B proteins from genotype 1b J4 and the 11 clinical isolates. All recombinant polymerases were active in the primer-dependent transcription assay (Table 2). We found that the affinity constants for both GTP and the primer/template were comparable to that of NS5B 1b J4. However, specific activities varied significantly among different enzyme isolates. While the 1b isolates had efficiencies similar to that of 1b J4, all other polymerases were less active due to differences in turnover numbers. Since all enzymes were purified in the same way and tested simultaneously in the polymerase assay, genotypic differences probably underlie these observations. Previous studies have already described possible variations in recombinant NS5B activity, caused not only by the method of production (15, 28) but also by minor changes in the amino acid sequence (27, 31).
FIG. 4.
SDS-PAGE analysis of NS5B enzyme isolates. Coomassie-stained 14% SDS-PAGE of purified recombinant enzymes. Lane M, BenchMark molecular mass marker (Invitrogen Carlsbad, CA). Electrospray mass spectrometry confirmed that all proteins had the predicted molecular masses (data not shown).
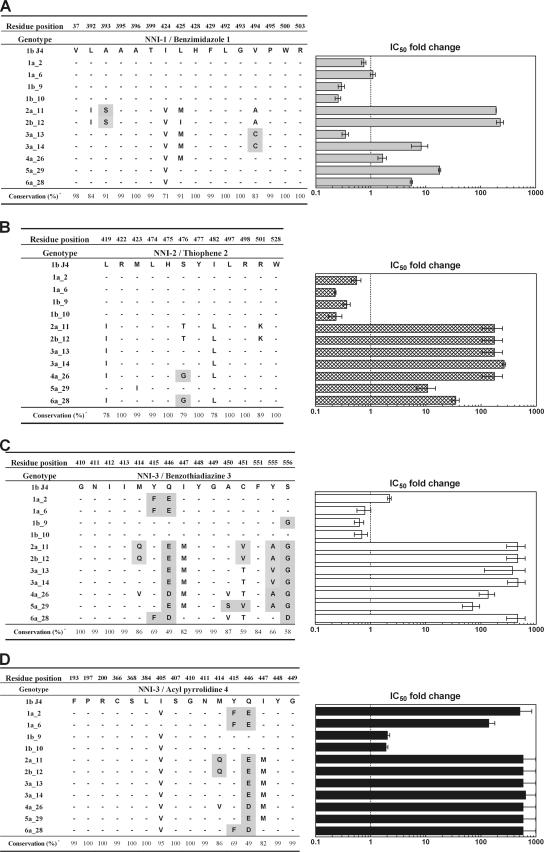
Next, we profiled the NNI reference inhibitors 1 to 3 and acyl pyrrolidine 4 on our NS5B genotypic panel. As for the NS5B mutant panel, the inhibitory effects measured for each enzyme isolate were reported as n-fold changes in IC50s relative to that for NS5B 1b J4. Based on the observed IC50 for 1b J4, we considered a compound with an IC50 n-fold change above 10-fold to be weakly active (>1 μM). Consistent with previous reports (P. L. Beaulieu, The identification of HCV polymerase inhibitors: a showcase of modern drug discovery, presented at the Medicinal Chemistry Gordon Research Conference, Colby-Sawyer College, New London, NH, 2005), we found that benzimidazole 1 maintained good inhibition across genotypes, except against 2a_11 and 2b_12, where it showed an approximate 100-fold increase in the IC50 (Fig. 5A). Thiophene 2 was able to inhibit only genotypes 1a, 1b, and 5a (Fig. 5B); this was unexpected, since structural analysis had predicted a versatile cross-genotypic coverage for this class of compound (55) and a close analog had been shown to inhibit genotype 2a polymerase (2). Similar to thiophene 2, benzothiadiazine 3 was capable of inhibiting genotypes 1a and 1b only (Fig. 5C); these findings are similar to what has been reported with a close analog (47). Unlike benzothiadiazine 3, we found that acyl pyrrolidine 4 inhibited only 1b_9 and 1b_10 polymerases, with a significant loss of activity for other genotypes, including genotype 1a (Fig. 5D).
FIG. 5.
Genotypic profiling of compounds 1 to 4. (A to D) Inhibitory activities of each compound are represented on the right panel next to the discontinuous determinants of its respective binding site. IC50 n-fold changes are reported relative to the IC50 of NS5BΔ21 1b J4. Determinants of NNI-1 and -2 are as previously reported (2, 13, 55). Determinants of NNI-3 are based on our docking model and a previous report (47). We do not exclude that residues other than these determinants might be involved for analogs or other compound classes. Nonconservative changes are shaded according to the following criteria: G, A, V, F, P, M, I, L, and W are nonpolar; S, T, C, Y, N, and Q are uncharged polar; and D, E, K, R, and H are charged polar. Conservative percentages from those residues believed to interact with the NNIs are obtained after aligning 467 NS5B sequences from different genotypes and subtypes (European HCV Database [http://euhcvdb.ibcp.fr/euHCVdb/]; accessed March 2007).
NNI-3 residues that affect genotypic coverage of acyl pyrrolidine 4 and benzothiadiazine 3.
To elucidate the NNI-3 binding site determinants (Fig. 5C and D) that cause the differences of inhibitory activity observed for acyl pyrrolidine 4 and benzothiadiazine 3 in genotypes 1 to 3, we mutated each position that varies in the enzyme isolates back to the wild-type 1b J4 sequence and tested whether the engineered mutations could restore the inhibitory activity observed with 1b J4 polymerase. For the acyl pyrrolidine, we generated 1b J4 (Y415A), 1a_6 (F415Y), and 1a_6 (F415Y/E446Q) to investigate the variations of genotype 1a. We found that F415Y considerably restored the inhibitory effect of acyl pyrrolidine 4 (Fig. 6A). These data indicate that Phe415 is the main cause of the loss of activity observed in genotype 1a and are consistent with the finding that no inhibition of 1b J4 (Y415A) was observed. Insertion of E446Q in the 1a_6 (F415Y) mutant resulted in a further modest increase of inhibition. However, the F415Y/E446Q double mutation was not able to restore full susceptibility to pyrrolidine inhibition relative to the level for 1b J4 (12-fold change relative to the level for 1b J4). Next, we investigated the variations of genotypes 2 and 3. Met414 is located in a lipophilic subpocket consisting of Pro197, Leu384, Met414, Tyr415, and Tyr448. The polar M414Q change of genotype 2a may disrupt hydrophobic contact. When acyl pyrrolidine 4 was tested against 2a_11 (Q414M), we found that Gln414 is not the main cause for the loss of inhibition in this genotype (Fig. 6B). Consistent with this, acyl pyrrolidine 4 also does not inhibit 3a_13 (identical binding site). We then checked for the importance of Glu446 and M447 and found that both M447I and E446Q could partially restore pyrrolidine inhibition (Fig. 6C, 580-fold and 920-fold change relative to the level for 1b J4, respectively, versus 1,080-fold for 2a_11). However, these effects are modest and do not explain the lack of activity observed with these genotypes. Glu446 and Met447 are both located in the β-flap, which is located next to the active site and is thought to be involved in de novo initiation (42). Superimposition of the 1b J4 NS5B structure with a 2a enzyme (2) revealed several important changes: (i) a 2-Å displacement of the β-flap; (ii) the rotation of Glu446; and (iii) the displacement of Met447 (Fig. 7). In silico analysis of each β-flap residue suggested that the presence of Phe445 in genotype 2a might trigger the observed conformational changes. To investigate this, we generated the 2a_11 (Q414M/F445C) double mutant but found that no inhibition could be restored, thereby ruling out Phe445. Finally, we docked acyl pyrrolidine 4 into the NNI-3 site of 1b J4 and observed that the β-flap wall becomes more shallow in genotype 2a and now clashes with the compound (Fig. 7). This suggests that repositioning of the β-flap wall may be the major cause for the loss of inhibition in genotype 2a and not the 414, 446, and/or 447 variations.
FIG. 6.
Inhibitory activities of compounds 3 and 4 on enzyme isolate mutants. Site-directed mutants of 1b J4, 1a_6, 2a_11, and 3a_13 were profiled against acyl pyrrolidine 4 (A and B) and benzothiadiazine 3 (C). In addition to the mutants shown, the following mutants were also generated and found not to be inhibited by these inhibitors (data not shown): 2a_11 (Q414M/V451T), 3a_13 (T451C), 3a_13 (V555Y), and 3a_13 (G556S).
FIG. 7.
Superimposition of the β-flap domain of genotypes 1b and 2a in the vicinity of acyl pyrrolidine 4. Stereo view of the β-flap and side chains that vary in genotypes 1b and 2a is illustrated in green for genotype 1b (PDB 1NB4) or in red for genotype 2a. The docking of acyl pyrrolidine 4 in genotype 1b was based on the structure of a proline sulfonamide analog (PDB 2GC8) (18). The represented β-flap wall is of genotype 2a. The structure of a closely related analog bound to a 1b BK enzyme was also used to dock acyl pyrrolidine 4 (PDB 2JC1) (data not shown). Both the inhibitor and the β-flap were found to overlap well with the model presented here. In addition to this, the observed β-flap displacement is always observed when all 1b and 2a enzymes available in the Protein Data Bank are superimposed.
Unlike acyl pyrrolidine 4, the capacity of benzothiadiazine 3 to inhibit genotype 2a could be partially restored with 2a_11 (Q414M). These data indicate that a polar change in the lipophilic subpocket is not tolerated by the benzothiadiazine analog. We then tested an extended panel of enzyme mutants of genotypes 2a and 3a and found that only the 3a_13 (M447I) mutant was able to restore modest benzothiadiazine inhibition (Fig. 6C). As for acyl pyrrolidine 4, these data do not explain the loss of activity observed with these genotypes and suggest that benzothiadiazine 3 may not be able to adapt to a more shallow β-flap wall.
DISCUSSION
Here we have shown that an NS5B enzyme panel comprising both NNI mutants and different genotypes can be used to address two key issues for the discovery of inhibitors, namely, the elucidation of the site of action and the profiling against variable genotypes. For the only reference inhibitor to which the binding site was not known, i.e., acyl pyrrolidine 4 (Burton et al., international patent application WO 2004009543 A2, 2004), clear evidence in support of NNI-3 was obtained. This finding is consistent with a previous report stating that the acyl pyrrolidines act as reversible, noncompetitive inhibitors with respect to GTP (5) and demonstrates the usefulness of our panel in the identification of hitherto unknown compound binding sites.
The poor clinical response to current therapy seen in patients infected with HCV genotype 1 has prompted drug development efforts to focus primarily on that genotype. Despite the better clinical response seen with genotypes 2 and 3, there also remains an unmet medical need for those populations, as well as for genotypes 4, 5, and 6. Changes in the incidence rates across genotypes may also impact future therapy: e.g., increases in the genotype 4 incidence in France have been well documented (39). Considering the most actively studied drug targets, it has been noted that both the protease and polymerase genes exhibit considerable diversity both across genotypes and at the quasispecies level (54). This has been studied most extensively for NS3, where the diversity has been characterized at the sequence (54, 57) and enzymatic levels and different sensitivities to protease inhibitors in development have been observed (48, 53). Similarly, in our NS5B drug discovery program, we wished to profile lead molecules for activity against a panel of clinically relevant genotypes, including multiple variants of genotypes 1a and 1b.
Using bulk sequencing of the predominant clone in each isolate, we derived sequences from >50 serum samples and selected those with sequences exhibiting divergence from the specific subtype consensus. Amino acid divergence across the genotypes was in the expected range (∼20 to 30%).
We observed differences in NS5B solubility, activity, and yield across genotypes and subtypes (data not shown). Such biophysical differences have been reported previously for 1a/1b recombinant polymerases (19). Nonetheless, this enzyme panel can be used to profile the activities of reference compounds. To better understand the differences observed across genotypes, we aligned the sequence of each NNI binding site of our enzyme isolates (based on the key residues identified by others) and examined whether the changes in amino acid side chains could account for the observed differences (Fig. 5). Consistent with previous reports (13, 55), changes in amino acid side chains were mainly conservative. However, we observed a nonconservative A393S change in the NNI-1 site. This change of an alanine to a serine may account for the loss of activity we observed when benzimidazole 1 was tested against 2a_11 and 2b_12 polymerases. In support of this, the X-ray structure of an NS5B/indole inhibitor complex (13) revealed that the indole binds to NNI-1 mainly through strong hydrophobic contacts of the cyclohexyl and phenyl ring, which fill two closely spaced pockets. Benzimidazole 1 is a close analog of the cocrystallized indole, and since our study and that of Di Marco et al. (13) used identical NS5B sequences in NNI-1, we speculate that benzimidazole 1 will also use the same moieties to contact the binding site. The A393S switch observed in genotype 2 introduces a polar group which potentially destabilizes the hydrophobic contact with the phenyl ring of the inhibitor. In addition, the V494A genotype 2a variation introduces a dimple in the pocket that may be deleterious to binding of the phenyl ring moiety.
Despite the high level of conservation of the NNI-2 pocket, we found that thiophene 2 could inhibit only the 1a/1b/5a enzyme isolates. This was somewhat unexpected, since two related compounds have been reported to bind in the NNI-2 site of a genotype 2a polymerase by X-ray crystallography and to be active inhibitors of the 1b J4 enzyme (2). These analogs, named inhibitor A and inhibitor B, differ from thiophene 2 at the methylcyclohexyl group by a 2,4-dichlorobenzoyl group and a 4-methylbenzoyl group, respectively. It was surprising to see the loss of activity of thiophene 2 on our 3a_13 and 3a_14 polymerases, since they harbor exactly the same NNI-2 residues as the genotype 2a enzyme reported above (2): Ile419 and Leu482 are swapped in genotype 3 relative to the 1b J4 sequence. These residues are involved in van der Waals interactions with the phenyl moiety present in thiophene 2 and in the reported structure of inhibitor A. This suggests that the benzoyl group of inhibitors A and B is involved in different interactions with NS5B than is the cyclohexyl moiety of thiophene 2, thereby compensating for the less-favorable 419/482 leucine/isoleucine swap. Because Ile419 and Leu482 are in close proximity to the phenyl moiety, the swapping of these residues slightly modifies the shape of the thiophene pocket and might destabilize the hydrophobic contact. Consistent with this, structural analysis of the NS5B/thiophene 2 bound complex reveals that the presence of the saturated cyclohexyl group perturbs the binding pocket and triggers structural changes that are not observed in the case of the nonsaturated benzoyl group (25). Interestingly, the Ile482Leu replicon mutant was found to be resistant to thiophene 2 in these studies (25).
Last, we found that NNI-3 was the least-conserved binding site in our genotypic panel. NNI-3 is the only pocket that exhibits subtype diversity between 1a and 1b. Interestingly, acyl pyrrolidine 4 inhibited only 1b_9 and 1b_10, unlike benzothiadiazine 3, which could inhibit all genotype 1 enzymes. We docked benzothiadiazine 3 in the NNI-3 binding site and compared our results with those in a previous report (47). Our model (data not shown) suggests a rationale for the loss of activity of benzothiadiazine against non-genotype-1 enzymes: a critical hydrogen bond mediated by Ser556 with the sulfonyl group of the inhibitor is lost by a change to Gly556, and a hydrophobic interaction between the inhibitor quinoline ring and Met414 is destabilized by a glutamine at this position. Consistent with this, some inhibition could be restored in the 2a_11 (Q414M) mutant. However, no inhibition could be observed with the 3a_13 (G556S) mutant, and 1b_9 is not affected by a glycine at this position. For acyl pyrrolidine, we found that Phe415 is crucial for genotype 1a coverage. The failure to restore full susceptibility to pyrrolidine inhibition on genotype 1a with the F415Y/E446Q double mutant suggests that the determinants of the acyl pyrrolidine binding site may extend beyond those presented in Fig. 5. For both NNI-3 inhibitors, we were not able to explain the loss of inhibition observed in genotypes 2/3 through the use of enzyme mutants. We propose that the bending of the β-flap wall may result in the loss of inhibition for these compounds. Further structural studies are needed to explore this hypothesis.
Our data illustrate the challenges of finding molecules capable of docking in pockets of enzymes from viruses with high genetic diversity, where single-residue changes can lead to a loss of inhibitor binding. From our analysis of the published structures, we conclude that benzimidazole 1, thiophene 2, and benzothiadiazine 3 contact NS5B mainly through hydrophobic interactions with the side chains. Hydrogen bonding and contacts with the polypeptide backbone are rare. To best adapt to highly variable binding sites, small molecules ideally use both enthalpy- and entropy-driven factors (37), i.e., the use of hydrogen bonding with the polypeptide backbone or highly conserved side chains and the use of flexible bonds that permit the hydrophobic groups to accommodate to diversity within the pool of binding sites, as has been shown with the human immunodeficiency virus protease inhibitor TMC114 (21). Thus, knowledge of a binding site across genotypes is a valuable tool for the rational design of future NS5B inhibitors.
ADDENDUM
The coordinates of the X-ray crystal structure of benzothiadiazine 3 and an acyl pyrrolidine analog in complex with NS5B (PDB 2FVC [47] and PDB 2JC1, respectively) were released during the revision of the manuscript and are in good agreement with our studies.
Acknowledgments
We thank Ina Vandenbroucke for technical assistance in the isolation of NS5B genes from clinical isolates. The pCV J4L6S plasmid was kindly provided by Jens Bukh of NIAID. We thank Reginald Clayton and Gregory Fanning for critical reading of the manuscript.
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Beaulieu, P. L., M. Bös, Y. Bousquet, P. DeRoy, G. Fazal, J. Gauthier, J. Gillard, S. Goulet, G. McKercher, M. A. Poupart, S. Valois, and G. Kukolj. 2004. Non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase: discovery of benzimidazole 5-carboxylic amide derivatives with low-nanomolar potency. Bioorg. Med. Chem. Lett. 14:967-971. [DOI] [PubMed] [Google Scholar]
- 2.Biswal, B. K., M. M. Cherney, M. Wang, L. Chan, C. G. Yannopoulos, D. Bilimoria, O. Nicolas, J. Bedard, and M. N. G. James. 2005. Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J. Biol. Chem. 280:18202-18210. [DOI] [PubMed] [Google Scholar]
- 3.Brass, V., H. E. Blum, and D. Moradpour. 2004. Recent developments in target identification against hepatitis C virus. Expert Opin. Ther. Targets 8:295-307. [DOI] [PubMed] [Google Scholar]
- 4.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, G., T. W. Ku, T. J. Carr, T. Kiesow, R. T. Sarisky, J. Lin-Goerke, A. Baker, D. L. Earnshaw, G. A. Hofmann, R. M. Keenan, and D. Dhanak. 2005. Identification of small molecule inhibitors of the hepatitis C virus RNA-dependent RNA polymerase from a pyrrolidine combinatorial mixture. Bioorg. Med. Chem. Lett. 15:1553-1556. [DOI] [PubMed] [Google Scholar]
- 6.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 7.Chan, L., O. Pereira, T. J. Reddy, S. K. Das, C. Poisson, M. Courchesne, M. Proulx, A. Siddiqui, C. G. Yannopoulos, N. Nguyen-Ba, C. Roy, D. Nasturica, C. Moinet, R. Bethell, M. Hamel, L. L'Heureux, M. David, O. Nicolas, P. Courtemanche-Asselin, S. Brunette, D. Bilimoria, and J. Bédard. 2004. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 2: tertiary amides. Bioorg. Med. Chem. Lett. 14:797-800. [DOI] [PubMed] [Google Scholar]
- 8.Chan, L., T. J. Reddy, M. Proulx, S. K. Das, O. Pereira, W. Wang, A. Siddiqui, C. G. Yannopoulos, C. Poisson, N. Turcotte, A. Drouin, M. H. Alaoui-Ismaili, R. Bethell, M. Hamel, L. L'Heureux, D. Bilimoria, and N. Nguyen-Ba. 2003. Identification of N,N-disubstituted phenylalanines as a novel class of inhibitors of hepatitis C NS5B polymerase. J. Med. Chem. 46:1283-1285. [DOI] [PubMed] [Google Scholar]
- 9.Condon, S. M., M. G. LaPorte, and T. Herbertz. 2005. Allosteric inhibitors of hepatitis C NS5B RNA-dependent RNA polymerase. Curr. Med. Chem. Anti-Infect. Agents 4:99-110. [Google Scholar]
- 10.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 11.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 12.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121-S127. [DOI] [PubMed] [Google Scholar]
- 13.Di Marco, S., C. Volpari, L. Tomei, S. Altamura, S. Harper, F. Narjes, U. Koch, M. Rowley, R. De Francesco, G. Migliaccio, and A. Carfi. 2005. Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. J. Biol. Chem. 280:29765-29770. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 15.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez, J., M. Martell, J. Quer, B. Cabot, and J. I. Esteban. 1999. Hepatitis C viral quasispecies. J. Viral Hepat. 6:3-16. [DOI] [PubMed] [Google Scholar]
- 17.Gopalsamy, A., K. Lim, G. Ciszewski, K. Park, J. W. Ellingboe, J. Bloom, S. Insaf, J. Upeslacis, T. S. Mansour, G. Krishnamurthy, M. Damarla, Y. Pyatski, D. Ho, A. Y. Howe, M. Orlowski, B. Feld, and J. O'Connell. 2004. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J. Med. Chem. 47:6603-6608. [DOI] [PubMed] [Google Scholar]
- 18.Gopalsamy, A., R. Chopra, K. Lim, G. Ciszewski, M. Shi, K. J. Curran, S. F. Sukits, K. Svenson, J. Bard, J. W. Ellingboe, A. Agarwal, G. Krishnamurthy, A. Y. M. Howe, M. Orlowski, B. Feld, J. O'Connell, and T. S. Mansour. 2006. Discovery of proline sulfonamides as potent and selective hepatitis C virus NS5b polymerase inhibitors. Evidence for a new NS5b polymerase binding site. J. Med. Chem. 49:3052-3055. [DOI] [PubMed] [Google Scholar]
- 19.Gu, B., L. L. Gutshall, D. Maley, C. M. Pruss, T. T. Nguyen, C. L. Silverman, J. Lin-Goerke, S. Khandekar, C. Liu, A. E. Baker, D. J. Casper, and R. T. Sarisky. 2004. Mapping cooperative activity of the hepatitis C virus RNA-dependent RNA polymerase using the genotype 1a-1b chimeras. Biochem. Biophys. Res. Commun. 313:343-350. [DOI] [PubMed] [Google Scholar]
- 20.Harper, S., B. Pacini, S. Avolio, M. Di Filippo, G. Migliaccio, R. Laufer, R. De Francesco, M. Rowley, and F. Narjes. 2005. Development and preliminary optimization of indole-N-acetamide inhibitors of hepatitis C virus NS5B polymerase. J. Med. Chem. 48:1314-1317. [DOI] [PubMed] [Google Scholar]
- 21.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, P. Wigerinck, M. P. de Béthune, and C. A. Schiffer. 2004. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 78:12012-12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong, L. B., L. B. Ye, L. Ye, K. A. Timani, Y. Zheng, Q. J. Liao, B. Z. Li, and B. Gao. 2006. Establishment of stable HeLa cell lines expressing enzymatically active hepatitis C virus RNA polymerase. Arch. Virol. 151:361-367. [DOI] [PubMed] [Google Scholar]
- 23.Kukolj, G., G. A. McGibbon, G. McKercher, M. Marquis, S. Lefebvre, L. Thavette, J. Gauthier, S. Goulet, M. A. Poupart, and P. L. Beaulieu. 2005. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J. Biol. Chem. 280:39260-39267. [DOI] [PubMed] [Google Scholar]
- 24.Lee, G., D. E. Piper, Z. Wang, J. Anzola, J. Powers, N. Walker, and Y. Li. 2006. Novel inhibitors of hepatitis C virus RNA-dependent RNA polymerase. J. Mol. Biol. 357:1051-1057. [DOI] [PubMed] [Google Scholar]
- 25.Le Pogam, S., H. Kang, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W. R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Nájera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 27.Lévêque, V. J. P., and Q. M. Wang. 2002. RNA-dependent RNA polymerase encoded by hepatitis C virus: biomedical applications. Cell. Mol. Life Sci. 59:909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludmerer, S. W., D. J. Graham, E. Boots, E. M. Murray, A. Simcoe, E. J. Markel, J. A. Grobler, O. A. Flores, D. B. Olsen, D. J. Hazuda, and R. L. LaFemina. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, H., V. Leveque, A. De Witte, W. Li, T. Hendricks, S. M. Clausen, N. Cammack, and K. Klumpp. 2005. Inhibition of native hepatitis C virus replicase by nucleotide and non-nucleoside inhibitors. Virology 332:8-15. [DOI] [PubMed] [Google Scholar]
- 33.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 34.Mo, H., L. Lu, T. Pilot-Matias, R. Pithawalla, R. Mondal, S. Masse, T. Dekhtyar, T. Ng, G. Koev, V. Stoll, K. D. Stewart, J. Pratt, P. Donner, T. Rockway, C. Maring, and A. Molla. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohno, T., and J. Y. N. Lau. 1996. The ‘gold-standard, ’ accuracy, and the current concepts: hepatitis C virus genotype and viremia. Hepatology 24:1312-1315. [DOI] [PubMed] [Google Scholar]
- 37.Ohtaka, H., and E. Freire. 2005. Adaptive inhibitors of the HIV-1 protease. Prog. Biophys. Mol. Biol. 88:193-208. [DOI] [PubMed] [Google Scholar]
- 38.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 39.Payan, C., F. Roudot-Thoraval, P. Marcellin, N. Bled, G. Duverlie, I. Fouchard-Hubert, P. Trimoulet, P. Couzigou, D. Cointe, C. Chaput, C. Henquell, A. Abergel, J. M. Pawlotsky, C. Hezode, M. Coude, A. Blanchi, S. Alain, V. Loustaud-Ratti, P. Chevallier, C. Trepo, V. Gerolami, I. Portal, P. Halfon, M. Bourliere, M. Bogard, E. Plouvier, C. Laffont, G. Agius, C. Silvain, V. Brodard, G. Thiefin, C. Buffet-Janvresse, G. Riachi, F. Grattard, T. Bourlet, F. Stoll-Keller, M. Doffoel, J. Izopet, K. Barange, M. Martinot-Peignoux, M. Branger, A. Rosenberg, P. Sogni, M. L. Chaix, S. Pol, V. Thibault, P. Opolon, A. Charrois, L. Serfaty, B. Fouqueray, J. D. Grange, J. J. Lefrere, and F. Lunel-Fabiani. 2005. Changing of hepatitis C virus genotype patterns in France at the beginning of the third millenium: the GEMHEP GenoCII Study. J. Viral Hepat. 12:405-413. [DOI] [PubMed] [Google Scholar]
- 40.Pfefferkorn, J. A., M. L. Greene, R. A. Nugent, R. J. Gross, M. A. Mitchell, B. C. Finzel, M. S. Harris, P. A. Wells, J. A. Shelly, R. A. Anstadt, R. E. Kilkuskie, L. A. Kopta, and F. J. Schwende. 2005. Inhibitors of HCV NS5B polymerase. Part 1: evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid. Bioorg. Med. Chem. Lett. 15:2481-2486. [DOI] [PubMed] [Google Scholar]
- 41.Pfefferkorn, J. A., R. Nugent, T. J. Groww, M. Greene, M. A. Mitchell, M. T. Reding, L. A. Funk, R. Anderson, P. A. Wells, J. A. Shelly, R. Anstadt, B. C. Finzel, M. S. Harris, R. E. Kilkuskie, L. A. Kopta, and F. J. Schwende. 2005. Inhibitors of HCV NS5B polymerase. Part 2: evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid. Bioorg. Med. Chem. Lett. 15:2812-2818. [DOI] [PubMed] [Google Scholar]
- 42.Powers, J. P., D. E. Piper, Y. Li, V. Mayorga, J. Anzola, J. M. Chen, J. C. Jaen, G. Lee, J. Liu, M. G. Peterson, G. R. Tonn, Q. Ye, N. P. Walker, and Z. Wang. 2006. SAR and mode of action of novel non-nucleoside inhibitors of hepatitis C NS5b RNA polymerase. J. Med. Chem. 49:1034-1046. [DOI] [PubMed] [Google Scholar]
- 43.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 44.Sarisky, R. T. 2004. Non-nucleoside inhibitors of the HCV polymerase. J. Antimicrob. Chemother. 54:14-16. [DOI] [PubMed] [Google Scholar]
- 45.Simmonds, P., J. Bukh, C. Combet, G. Deléage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspé, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, T. Shin-I, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 46.Tan, S. L., A. Pause, Y. Shi, and N. Sonenberg. 2002. Hepatitis C therapeutics: current status and emerging strategies. Nat. Rev. Drug Discov. 1:867-881. [DOI] [PubMed] [Google Scholar]
- 47.Tedesco, R., A. N. Shaw, R. Bambal, D. Chai, N. O. Concha, M. G. Darcy, D. Dhanak, D. M. Fitch, A. Gates, W. G. Gerhardt, D. L. Halegoua, C. Han, G. A. Hofmann, V. K. Johnston, A. C Kaura, N. Liu, R. M. Keenan, J. Lin-Goerke, R. T. Sarisky, K. J. Wiggall, M. N. Zimmerman, and K. J. Duffy. 2006. 3-(1,1-Dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem. 49:971-983. [DOI] [PubMed] [Google Scholar]
- 48.Thibeault, D., C. Bousquet, R. Gingras, L. Lagace, R. Maurice, P. W. White, and D. Lamarre. 2004. Sensitivity of NS3 serine proteases from hepatitis C virus genotypes 2 and 3 to the inhibitor BILN 2061. J. Virol. 78:7352-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomei, L., S. Altamura, G. Paonessa, R. De Francesco, and G. Migliaccio. 2005. HCV antiviral resistance: the impact of in vitro studies on the development of antiviral agents targeting the viral NS5B polymerase. Antivir. Chem. Chemother. 16:225-245. [DOI] [PubMed] [Google Scholar]
- 51.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. De Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong, X., Z. Guo, J. Wright-Minogue, E. Xia, A. Prongay, V. Madison, P. Qiu, S. Venkatraman, F. Velazquez, F. G. Njoroge, and B. A. Malcolm. 2006. Impact of naturally occurring variants of HCV protease on the binding of different classes of protease inhibitors. Biochemistry 45:1353-1361. [DOI] [PubMed] [Google Scholar]
- 54.Vallet, S., S. Gouriou, J. B. Nousbaum, M. C. Legrand-Quillien, A. Goudeau, and B. Picard. 2005. Genetic heterogeneity of the NS3 protease gene in hepatitis C virus genotype 1 from untreated infected patients. J. Med. Virol. 75:528-537. [DOI] [PubMed] [Google Scholar]
- 55.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. G. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 56.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 57.Winters, M. A., S. L. Welles, and M. Holodniy. 2006. Hepatitis C virus protease gene diversity in patients coinfected with human immunodeficiency virus. J. Virol. 80:4196-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]