Abstract
Clinical isolates of measles virus (MV) use signaling lymphocyte activation molecule (SLAM) as a cellular receptor, whereas vaccine and laboratory strains may utilize the ubiquitously expressed CD46 as an additional receptor. MVs also infect, albeit inefficiently, SLAM− cells, via a SLAM- and CD46-independent pathway. Our previous study with recombinant chimeric viruses revealed that not only the receptor-binding hemagglutinin (H) but also the matrix (M) protein of the Edmonston vaccine strain can confer on an MV clinical isolate the ability to grow well in SLAM− Vero cells. Two substitutions (P64S and E89K) in the M protein which are present in many vaccine strains were found to be responsible for the efficient growth of recombinant virus in Vero cells. Here we show that the P64S and E89K substitutions allow a strong interaction of the M protein with the cytoplasmic tail of the H protein, thereby enhancing the assembly of infectious particles in Vero cells. These substitutions, however, are not necessarily advantageous for MVs, as they inhibit SLAM-dependent cell-cell fusion, thus reducing virus growth in SLAM+ B-lymphoblastoid B95a cells. When the cytoplasmic tail of the H protein is deleted, a virus with an M protein possessing the P64S and E89K substitutions no longer grows well in Vero cells yet causes cell-cell fusion and replicates efficiently in B95a cells. These results reveal a novel mechanism of adaptation and attenuation of MV in which the altered interaction of the M protein with the cytoplasmic tail of the H protein modulates MV growth in different cell types.
Measles is an acute contagious disease characterized by high fever and a typical rash. Despite the availability of effective live vaccines, ∼30 million cases and approximately half a million deaths related to measles are reported each year worldwide (12). Measles virus (MV), the causative agent of measles, belongs to the genus Morbillivirus in the family Paramyxoviridae and has a nonsegmented, negative-sense RNA genome of ∼16,000 nucleotides. The genome is encapsidated by the nucleocapsid (N) protein and is associated with a viral RNA-dependent RNA polymerase composed of two subunits, the phospho (P)- and large (L) proteins, forming a helical ribonucleoprotein complex (RNP). Each virion has glycoprotein spikes, the hemagglutinin (H) and fusion (F) proteins, on the envelope and attaches to cells via binding of the H protein to cellular receptors (12). Signaling lymphocyte activation molecule (SLAM; also known as CD150) is a receptor for clinical isolates of MV, while vaccine strains of MV use the ubiquitously expressed CD46 as a receptor as well as SLAM (53, 59). Binding of the H protein to a receptor triggers the process of fusion of the virus envelope with the plasma membrane, which is mediated by the F protein (57). RNP is released into the cytoplasm and initiates the transcription of viral mRNAs. As newly synthesized viral proteins accumulate, the same RNP is used as a template for replication of the virus genome via synthesis of a positive-strand antigenome intermediate. MV then spreads in cell cultures or animal tissues in two ways, either via the production of progeny virus particles that undergo successive rounds of infection or by fusion of infected cells with neighboring uninfected cells (cell-cell fusion). The matrix (M) protein plays a crucial role in the assembly of progeny virus particles by interacting with the RNP (15) and the cytoplasmic tails of the H and F proteins (4, 5, 43), whereas expression of the H and F proteins alone is sufficient to induce cell-cell fusion (57).
MV strains isolated from B-lymphoid cell lines reproduce a clinical course of measles in experimentally infected monkeys (20, 21). These virulent MV strains use SLAM but not CD46 as a cellular receptor (60). Since SLAM is expressed only on cells of the immune system, virulent MV strains enter nonlymphoid cells inefficiently and fail to cause cell-cell fusion in them. Accordingly, virulent MV strains hardly replicate in nonlymphoid cells. In contrast, vaccine strains of MV, obtained by numerous rounds of passage of the original isolate in various cultured cells, grow efficiently in nonlymphoid cells, as H proteins of vaccine strains have the ability to bind the ubiquitously expressed CD46 (60). The vaccine strains of MV are safe and very effective, but the molecular bases of their attenuation and efficacy are poorly understood.
In our previous study, recombinant chimeric MVs in which part of the genome of the virulent IC-B strain was replaced with the corresponding sequences from the Edmonston vaccine strain were generated (45). The parental IC-B strain could not grow in Vero cells (CD46+ SLAM−) derived from monkey kidneys. Upon replacement of the M gene alone with that of the Edmonston strain, the recombinant virus replicated efficiently in Vero cells, although it still entered cells inefficiently (45). The recombinant virus apparently infected Vero cells via a SLAM- and CD46-independent mechanism of entry (13). P64S and E89K substitutions were shown to be responsible for the ability of the Edmonston M protein to enable the virus to grow efficiently in Vero cells at postentry steps (45). Examination of published sequence data revealed that all MV vaccine strains have either or both of the P64S and E89K substitutions in their M proteins (32, 38, 39) (GenBank accession number EF033071).
In this study, we show that the M protein with the P64S and E89K substitutions (hereafter called the M-P64S/E89K protein) largely colocalizes with the H protein in cells in the absence of other viral components, unlike the wild-type (wt) M protein (hereafter called the M-wt protein). Furthermore, the M-P64S/E89K protein enhances the assembly and release of infectious virus particles, contributing to efficient growth in SLAM− nonlymphoid cells. However, these substitutions are not necessarily advantageous for MV growth, as they strongly inhibit SLAM-mediated cell-cell fusion and thereby retard virus growth in SLAM+ lymphoid cells. Thus, these substitutions may partly account for the attenuation of MV vaccine strains. Defects in virus growth and cell-cell fusion in SLAM+ cells caused by the P64S and E89K substitutions in the M protein can be relieved by truncating the cytoplasmic tail of the H protein, even though the virus loses the ability to replicate in Vero cells. Our data suggest that MV can optimize the mode of virus spread (cell-cell fusion or infectious particle production) by acquiring these substitutions in the M protein, thereby modulating its interaction with the cytoplasmic tail of the H protein.
MATERIALS AND METHODS
Cells and viruses.
Vero cells constitutively expressing human SLAM (Vero/hSLAM) (31) were maintained in Dulbecco's modified Eagle's medium (DMEM; ICN Biomedicals, Aurora, OH) supplemented with 7.5% fetal bovine serum (FBS) and 500 μg/ml Geneticin (G418; Nacalai Tesque, Tokyo, Japan). CHO cells constitutively expressing human SLAM (CHO/hSLAM) (53) were maintained in RPMI medium (ICN Biomedicals) supplemented with 7.5% FBS and 500 μg/ml G418. B95a (20) and CHO cells were maintained in RPMI medium supplemented with 7.5% FBS. Vero and HeLa cells were maintained in DMEM supplemented with 7.5% FBS. Recombinant MVs were generated from cDNAs by using CHO/hSLAM cells and a vaccinia virus expressing T7 RNA polymerase (vTF7-3; a gift from B. Moss) (10) or LO-T7-1 (19, 61) (a gift from M. Kohara), as reported previously (27, 47). The generated MVs were propagated in B95a cells, and virus stocks after two or three passages in B95a cells were used in further experiments.
Plasmid construction.
All full-length genome plasmids were derived from the p(+)MV323 plasmid, which encodes the antigenomic full-length cDNA of the wt IC-B strain of MV (49). The p(+)MV323-EGFP plasmid, which has an additional transcriptional unit encoding enhanced green fluorescent protein (EGFP), was reported previously (13). The plasmids p(+)MV323-EGFP-M/P64S/E89K and -M/P64S/E89K/A209T were reported previously (45). Plasmids p(+)MV323-EGFP-HΔ20 and p(+)MV323-EGFP-HΔ20-M/P64S/E89K were constructed as follows. PCR was performed using p(+)MV323-EGFP as the template and a specific primer pair (5′-TCTTAATTAAAACTTAGGGTGCAAGATCATCCACAATGAGGATAGTTATTAACAGAGAA-3′ and 5′-ACACTAGTGGGTATGCCTGATGTCTGGGTGACATCATGTGATCGGTTCACTGGCAGCCCTATCTGCGATTGGTTCCATC-3′). The generated DNA fragment was digested with PacI and SpeI, and the released fragment was used to replace the PacI-SpeI fragment (nucleotide positions 7,238 to 9,175) of the p(+)MV323-EGFP or p(+)MV-EGFP-M/P64S/E89K plasmid (nucleotide position numbers are shown in accordance with the sequence of the IC-B strain genome [50]). Plasmids p(+)MV323-EGFP-FΔ30 and p(+)MV323-EGFP-FΔ30-M/P64S/E89K were constructed by introducing three tandem stop codons into the p(+)MV323-EGFP and p(+)MV-EGFP-M/P64S/E89K plasmids, respectively, by site-directed mutagenesis using a complementary primer pair (5′-GCAGGGGGCGCTAGTAGTAAAAGGGAGAA-3′ and 5′-TTCTCCCTTTTACTACTAGCGCCCCCTGC-3′). The coding regions for the wt and mutated M, HΔ20, and FΔ30 proteins were cloned into the eukaryotic expression plasmid pCA7 (48), a derivative of pCAGGS (29). The pCA7-ICH and pCA7-ICF plasmids, encoding the wt H and F proteins, respectively, were reported previously (44).
Syncytium formation assay by phase-contrast microscopy.
B95a cells cultured in six-well culture plates were infected with vTF7-3 at a multiplicity of infection (MOI) of 0.5, incubated for 1 h at 37°C, and then cotransfected with pCA7-ICH (1 μg) and pCA7-ICF (1 μg) together with either empty pCA7 vector or that encoding the M-wt or M-P64S/E89K protein (pCA7-ICM and pCA7-MP64S/E89K, respectively), using the cationic lipid transfection reagent Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. The cells were infected with vTF7-3 to enhance expression from the plasmids containing the T7 promoter. At 24 h posttransfection, the transfected cells were observed under a phase-contrast imaging microscope.
Quantitative fusion assay.
A quantitative fusion assay was performed using a method described previously (30, 44), with minor modifications. Briefly, monolayers of Vero cells (effector cells) in 24-well cluster plates were infected with MVA-T7 (58) (a gift from B. Moss) at an MOI of 0.5, incubated for 1 h at 37°C, and then transfected with 0.2 μg each of an H protein-encoding plasmid (pCA7-ICH or pCA7-ICHΔ20), an F protein-encoding plasmid (pCA7-ICF or pCA7-ICFΔ30), and an M protein-encoding plasmid (pCA7-ICM or pCA7 encoding each mutated M protein) per well, using the Lipofectamine 2000 reagent (Invitrogen). Monolayers of B95a cells (target cells) in 24-well cluster plates were transfected with 0.5 μg of pG1NT7βgal (30) (a gift from E. A. Berger), a plasmid containing the lacZ gene under the control of the T7 promoter. Twelve hours after transfection, target cells were harvested, resuspended in RPMI medium with 7.5% FBS, and transferred onto the monolayers of effector cells. After 7 h of incubation, β-galactosidase activity in cells was analyzed by a β-galactosidase reporter gene assay, using a chemiluminescence kit (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. Chemiluminescence was measured using a Mithras LB940 plate reader (Berthold Technologies, Pforzheim, Germany).
Virus titration.
Monolayers of Vero/hSLAM cells in 24-well cluster plates were infected with 50 μl of serially diluted virus samples and incubated for 1 h at 37°C. After 1 h of incubation, 150 μl of DMEM supplemented with 7.5% FBS and 100 μg/ml of a fusion block peptide (Z-D-Phe-Phe-Gly; Peptide Institute Inc., Osaka, Japan) (37) was added to each well to block the second round of infection by progeny viruses. At 36 h postinfection (p.i.), the number of EGFP-expressing cells was counted under a fluorescence microscope. The number was expressed in cell infectious units (CIU). CIU are essentially comparable to PFU (48).
Surface biotinylation, immunoprecipitation, and Western blot analysis.
At 48 h p.i., infected B95a cells were washed three times with cold phosphate-buffered saline (PBS) containing 0.1 mM CaCl2 and 1 mM MgCl2 and incubated twice for 30 min each at 4°C with 2 mg of sulfo-N-hydroxysuccinimidobiotin (Funakoshi, Tokyo, Japan) per ml. After that, the cells were washed once with cold PBS containing 0.1 M glycine and three times with cold PBS. Cells were lysed in 0.5 ml of radioimmunoprecipitation assay (RIPA) buffer (0.15 mM NaCl, 0.05 mM Tris-HCl, pH 7.2, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) and then centrifuged for 40 min at 15,000 rpm. For immunoprecipitation, an MV H protein-specific monoclonal antibody (MAb) (E396; a gift from T. A. Sato) or an MV F protein-specific MAb (E12; a gift from T. A. Sato) was added to samples and incubated for 16 h at 4°C. Immune complexes were precipitated by incubation with protein A and G Sepharose (1:1 mixture) (Amersham Biosciences, Piscataway, NJ) for 30 min at 4°C. Sepharose beads were washed three times with 1× RIPA buffer plus 0.3 M NaCl, once with 1× RIPA buffer plus 0.15 M NaCl, and once with SDS wash II buffer (150 mM NaCl, 50 mM Tris, pH 7.4, 2.5 mM EDTA) (33). Samples were resuspended in SDS loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min, and polypeptides were fractionated by SDS-polyacrylamide gel electrophoresis on a 10% polyacrylamide gel. Polypeptides were then blotted onto a polyvinylidene difluoride membrane (Hybond-P; Amersham Biosciences), using the semidry blot technique. Biotinylated H and F proteins were detected by incubation with horseradish peroxidase-conjugated streptavidin (Zymed Laboratories, San Francisco, CA). Total H and F proteins were detected using rabbit sera against the H protein (a gift from T. Kohama) and the F protein (5) (a gift from R. Cattaneo), with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Amersham Biosciences) as the secondary antibody. The membranes were treated with ECL Plus reagent (Amersham Biosciences), and chemiluminescence signals on the membranes were detected and visualized using a VersaDoc 3000 imager (Bio-Rad, Hercules, CA).
Assay for synthesis of viral mRNAs, genomes, and proteins and for production of infectious particles.
Monolayers of Vero/hSLAM cells in six-well cluster plates were infected with recombinant MVs at an MOI of 1.0 per cell and then incubated for 3 h at 37°C. After 3 h of incubation, the cells were washed with PBS and incubated in 1 ml of DMEM containing 7.5% FBS and 10 μg of an anti-human SLAM MAb (IPO-3; Kamiya Biomedical, Seattle, WA) that blocks the second round of infection and cell-to-cell fusion. At 48 h p.i., levels of viral RNAs and proteins, as well as production of infectious particles, were analyzed. Viral mRNAs and genomes were quantified by reverse transcription-quantitative RCR (RT-QPCR), as reported previously (28, 48). Viral proteins (N, P, V, M, and H) were detected by immunoblotting and quantified using a VersaDoc 3000 imager (Bio-Rad), as reported previously (28, 48). Cell-free virus was obtained from the culture supernatants, and cell-associated virus was recovered from infected cells by sonication. The CIU was determined on Vero/hSLAM cells.
Immunofluorescence staining.
HeLa cells were seeded on coverslips in six-well plates and transfected with pCA7 plasmids encoding the wt and mutated H, F, and M proteins, using the Lipofectamine 2000 reagent (Invitrogen). At 48 h posttransfection, cells were fixed and permeabilized with PBS containing 2.5% formaldehyde and 0.5% Triton X-100. Fixed cells were washed with PBS and incubated with a primary MAb against the H (E396), F (E12), or M (E388; a gift from T. A. Sato) protein and with a rabbit antiserum raised against the H or M protein (a gift from T. Kohama) for 1 h at 37°C, followed by incubation with Alexa Fluor 488-conjugated and Alexa Fluor 594-conjugated secondary antibodies (Molecular Probes, Eugene, OR) for 1 h at 37°C. The cells were observed using a confocal microscope (Radiance 2100; Bio-Rad).
RESULTS
P64S and E89K substitutions in the M protein enhance the assembly and release of infectious particles.
Our previous study indicated that P64S and E89K substitutions in the M protein conferred on the virulent IC-B strain of MV the ability to replicate efficiently in Vero cells, although the mutant virus still entered cells inefficiently, like the parental virus (45). To understand the mechanism by which the M-P64S/E89K protein promotes MV growth in Vero cells at postentry steps, the replication abilities of the IC323-EGFP virus with the M-wt protein (hereafter called the wt virus) and that with the M-P64S/E89K protein (hereafter called the P64S/E89K virus) were analyzed. IC323-EGFP is a recombinant MV based on the IC-B strain with an additional expression unit for EGFP (13).
Since both the wt and P64S/E89K viruses entered Vero cells inefficiently, it was difficult to perform single-step growth analysis of these viruses in Vero cells. Therefore, Vero/hSLAM cells, which allow efficient virus entry, were used for the analysis. To block the second round of infection, anti-human SLAM MAb (IPO-3) was added to the culture medium at 3 h p.i. At 48 h p.i., infectious titers in culture medium (cell-free) and cells (cell-associated) were analyzed. Cell-free and cell-associated titers of P64S/E89K virus were ∼200 and ∼30 times higher, respectively, than those of wt virus (Fig. 1A). Cell-free and cell-associated virus titers of the P64S/E89K virus in Vero cells at 48 h p.i. were ∼102 and ∼103 CIU/ml, respectively, whereas those of wt virus were negligible. Immunoblot analysis and RT-QPCR showed that the amounts of viral proteins and RNAs synthesized in Vero/hSLAM cells infected with these two viruses were comparable at 48 h p.i. (Fig. 1B and C). Thus, the results indicate that the P64S and E89K substitutions in the M protein promote the assembly and release of MV particles in Vero/hSLAM cells (and presumably also in Vero cells).
FIG. 1.
P64S and E89K substitutions in the M protein increase the production of infectious particles. (A) Vero/hSLAM cells were infected with wt and P64S/E89K viruses at an MOI of 1.0; 3 h after infection, an anti-human SLAM antibody (IPO-3) was added to block the second round of infection and cell-to-cell fusion. Infectious virus titers in culture medium (cell-free) and cells (cell-associated) were determined in Vero/hSLAM cells 48 h after infection. White bars, wt virus; gray bars, P64S/E89K virus. (B) Forty-eight hours after infection, the N, P, V, M, and H proteins were detected in virus-infected cells by immunoblotting. (C) Forty-eight hours after infection, levels of viral mRNAs and genomes were analyzed by RT-QPCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. White bars, wt virus; gray bars, P64S/E89K virus.
The M-P64S/E89K protein inhibits SLAM-dependent cell-cell fusion.
It was notable that the P64S/E89K virus hardly produced syncytia in SLAM+ B95a cells, unlike the wt virus (Fig. 2A). To better understand the mechanism by which the replacement of the M-wt protein with the M-P64S/E89K protein affected the ability of the wt virus to induce cell-cell fusion, the H and F proteins were expressed on B95a cells, using expression plasmids, together with the M-wt or M-P64S/E89K protein. Expression of the H and F proteins alone produced large syncytia in B95a cells (Fig. 2B, H+F), and coexpression of the M-wt protein enhanced syncytium formation (Fig. 2B, H+F+M-wt). In contrast, when the M-P64S/E89K protein was coexpressed, syncytium formation was greatly restricted (Fig. 2B, H+F+M-P64S/E89K). In all cases, the anti-SLAM MAb IPO-3 completely blocked syncytium formation (data not shown). Thus, whereas the MV-wt protein promoted SLAM-dependent cell-cell fusion induced by the H and F proteins, the M-P64S/E89K protein inhibited this process.
FIG. 2.
P64S and E89K substitutions in the M protein inhibit cell-cell fusion in B95a cells. (A) Syncytium formation by recombinant MVs. B95a cells were infected with the wt or P64S/E89K virus at an MOI of 0.01. At 48 h p.i., they were observed under a phase-contrast and a fluorescence microscope. Magnification, ×100. (B) Syncytium formation by expression plasmids. B95a cells infected with vTF7-3 were cotransfected with H and F protein expression plasmids (H+F). They were also cotransfected with the M-wt or M-P64S/E89K protein expression plasmid together with the H and F protein expression plasmids (H+F+M-wt and H+F+M-P64S/E89K, respectively). At 24 h posttransfection, the cells were observed under a phase-contrast microscope. Magnification, ×100. (C) Expression levels of glycoproteins of wt and P64S/E89K viruses. B95a cells were infected with the wt or P64S/E89K virus. At 48 h p.i., the cells were incubated with biotin and then lysed. The H and F proteins were immunoprecipitated with H and F protein-specific MAbs, subjected to SDS-polyacrylamide gel electrophoresis, and blotted onto polyvinylidene difluoride membranes. One membrane was incubated with streptavidin-conjugated peroxidase to detect biotinylated H and F proteins (surface). Another membrane was incubated with rabbit antisera raised against the H and F proteins and peroxidase-conjugated anti-rabbit immunoglobulins to detect the total amounts of H and F proteins.
In the above experiment, the M-P64S/E89K protein may have inhibited syncytium formation by reducing expression of the H and/or F protein on the cell surface. To examine this possibility, the amounts of H and F proteins in B95a cells infected with the wt or P64S/E89K virus were analyzed. The amounts of biotinylated H and F proteins expressed on the cell surface, as well as the total amounts of H and F proteins, were comparable in cells infected with either virus (Fig. 2C). Flow cytometry analysis also confirmed that the cell surface expression of the H protein was comparable in these cells (data not shown). These results indicate that the M-P64S/E89K protein did not affect the expression levels of H and F proteins on the cell surface but directly affected the fusion process.
Strong colocalization of the M-P64S/E89K protein with the H protein.
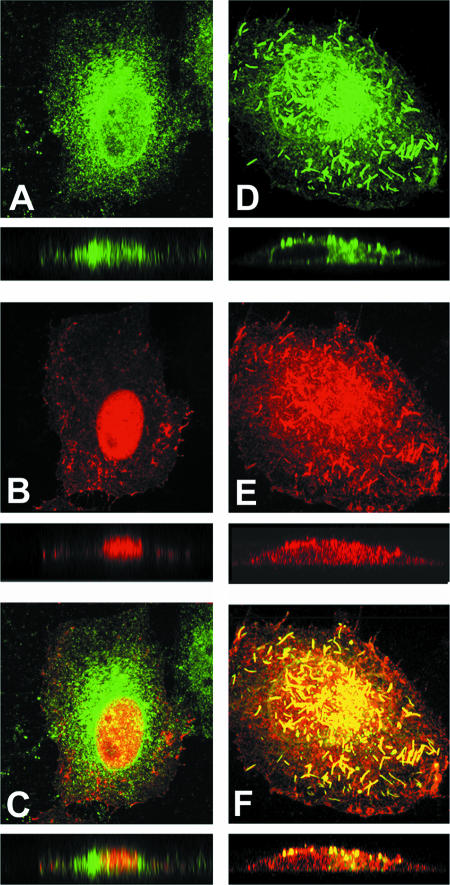
Cathomen et al. (5) showed with the MV Edmonston vaccine strain that the interaction of the M protein with the cytoplasmic tails of the viral envelope glycoproteins strongly affects cell-cell fusion. To examine the ability of the M-wt and M-P64S/E89K proteins to interact with the H and F proteins, the distributions of these viral proteins in cells were analyzed by confocal microscopy. When expressed alone, the F and H proteins localized in the perinuclear region of the cells, presumably the Golgi apparatus, as well as at the cell surface, showing a punctate staining pattern (data not shown). The M protein (either the M-wt or M-P64S/E89K protein) was distributed both in the nucleus and in the cytoplasm, but it was predominantly found in the nucleus (Fig. 3A and E). When coexpressed with the F protein, the M-wt protein partly colocalized with it, showing a punctate staining pattern (data not shown). In contrast, the M-wt protein did not colocalize with the H protein (Fig. 3B). Coexpression of the M-wt, H, and F proteins did not show an apparent redistribution of these viral proteins, and the M-wt protein still hardly colocalized with the H protein (Fig. 3C). Coexpression of the N protein with the M-wt, H, and F proteins did not alter the distributions of the latter proteins (data not shown). However, when coexpressed with the H, F, N, and P proteins, the M-wt protein redistributed and strongly colocalized not only with the F protein but also with the H protein (Fig. 3D). These results suggest that the P protein of MV plays an important role in virus assembly. Schmitt et al. (42) showed with parainfluenza virus 5 that expression of N, M, and either glycoprotein (F or hemagglutinin-neuraminidase [HN]) is required for efficient virus assembly and budding. Coronel et al. (7) reported that nucleocapsid incorporation into human parainfluenza virus 1 is regulated by a specific interaction between the N and M proteins. Since the P protein of MV has the ability to retain the N protein in the cytoplasm (16), it may thereby indirectly promote the coalescence of the M-wt, H, and N proteins.
FIG. 3.
Distributions of viral proteins analyzed by indirect immunofluorescence and confocal microscopy. MV proteins (H, F, N, P, M-wt, and M-P64S/E89K proteins) were expressed in HeLa cells in various combinations, using expression plasmids. (A) M-wt; (B) M-wt and H; (C) M-wt, H, and F; (D) M-wt, H, F, N, and P; (E) M-P64S/E89K; (F) M-P64S/E89K and H; (G) M-P64S/E89K, H, and F; (H) M-P64S/E89K, H, F, N, and P. At 48 h posttransfection, cells were permeabilized, fixed, and incubated with a MAb (E388) specific to the M protein and with rabbit antiserum against the H protein, followed by incubation with Alexa Fluor 594-conjugated anti-mouse and Alexa Fluor 488-conjugated anti-rabbit secondary antibodies. x-y plane images of 30 compiled sections are shown in the top panels. z-Scan images of 30 compiled sections are shown below the x-y plane images. Dual fluorescence images are shown. Magnification, ×600. The boundaries of the nuclei are shown with dashed lines (D and H).
In contrast, when the H, F, and M-P64S/E89K proteins were expressed together, they mostly colocalized in brightly stained filamentous structures, even in the absence of the N and P proteins (Fig. 3G). The z plane reconstruction showed that the filamentous structures were formed at the cell surface (Fig. 3G). They formed efficiently even when only the H protein was coexpressed with the M-P64S/E89K protein (Fig. 3F). Coexpression of the M-P64S/E89K and F proteins in the absence of the H protein also resulted in the formation of filamentous structures, albeit to a much lesser extent (data not shown). The filamentous structures became much less evident when the N and P proteins were coexpressed (Fig. 3H). Since electron microscopy analyses showed that virions of the P64S/E89K virus were pleomorphic (but typically were spherical particles) and indistinguishable in morphology from the wt virus particles (data not shown), the presence of the N and P proteins may have allowed the P64S/E89K virus to form spherical, but not filamentous, particles.
An H protein with a truncated cytoplasmic tail no longer colocalizes with the M-P64S/E89K protein.
We speculated that the interaction between the M-P64S/E89K protein and the cytoplasmic tail of the H protein may have enhanced virus assembly and inhibited cell-cell fusion. Moll et al. (25) showed that when the cytoplasmic tails of the H and F proteins of the MV Edmonston strain were truncated by 20 and 30 amino acids, respectively, the proteins were expressed normally on the cell surface but lost the ability to interact with the M protein. To confirm the importance of the interaction between the M and H proteins in our findings, expression plasmids were generated that encoded the H and F proteins of the IC-B strain, with 20- and 30-amino-acid truncations, respectively, in their cytoplasmic tails (HΔ20 and FΔ30). When expressed alone, both HΔ20 and FΔ30 localized to the perinuclear region and showed a punctate staining pattern at the cell surface, like the intact H and F proteins (H-wt and F-wt, respectively), consistent with the data by Moll et al. (25). When the HΔ20 and FΔ30 proteins were coexpressed with the M-P64S/E89K protein, they were distributed differently from the H-wt and F-wt proteins. The HΔ20 protein neither colocalized with the M-P64S/E89K protein nor formed filamentous structures (Fig. 4A to C). Since no antibody reacting with the ectodomain of the F protein was available, the FΔ30 protein could not be detected in indirect immunofluorescence assays. Cell surface expression of the FΔ30 protein was confirmed by a functional cell-cell fusion assay (Fig. 5). Coexpression of FΔ30 with the M-P64S/E89K and H-wt proteins did not affect the colocalization of those two proteins or the formation of filamentous structures (Fig. 4D to F). These data indicate that the cytoplasmic tail of the H protein of the IC-B strain plays a crucial role in the interaction with the M-P64S/E89K protein as well as in the formation of filamentous structures.
FIG. 4.
Effect of deletion of the cytoplasmic tail of the H or F protein on the distribution of MV proteins. M-P64S/E89K, F-wt, and HΔ20 proteins (A to C) or M-P64S/E89K, FΔ30, and H-wt proteins (D to F) were coexpressed in HeLa cells by using expression plasmids. At 48 h posttransfection, cells were permeabilized, fixed, and incubated with a MAb (E388) specific to the M protein (B and E) or with rabbit antiserum against the H protein (A and D), followed by incubation with Alexa Fluor 594-conjugated anti-mouse (B and E) and Alexa Fluor 488-conjugated anti-rabbit (A and D) secondary antibodies. Single (A, B, D, and E) and dual (C and F) fluorescence images are shown. Magnification, ×600.
FIG. 5.
Effect of deletion of the cytoplasmic tail of the H or F protein on cell-cell fusion. (A) Quantification of cell-cell fusion. B95a cells transfected with pG1NT7βgal were mixed with MVA-T7-infected Vero cells expressing the F, H, and M proteins. After 7 h of incubation, cell-cell fusion was quantified by measuring β-galactosidase activity. β-Galactosidase activity in Vero cells expressing the H and F proteins but not the M protein was set to 100%. β-Galactosidase activity in Vero cells expressing the F and M proteins but not the H protein was set to 0%. The bars indicate the means ± standard deviations for triplicate samples. (B) Syncytium formation in MV-infected cells. B95a cells were infected with recombinant MVs at an MOI of 0.01. Panels show representative images obtained with a phase-contrast and a fluorescence microscope 2 days after infection. Magnification, ×100.
Effects of deletion of the cytoplasmic tails of the H and F proteins on cell-cell fusion.
The M-P64S/E89K protein inhibited cell-cell fusion mediated by the H and F proteins, whereas the M-wt protein enhanced this process (Fig. 2). The levels of cell-cell fusion were quantified using a previously described method (44) and are shown in Fig. 5A. The cell-cell fusion activities of the H-wt and F-wt proteins in the absence of M protein were set to 100%. Coexpression of the M-wt protein with the H-wt and F-wt proteins increased the activity to ∼150%. In contrast, coexpression of the M-P64S/E89K protein with the H-wt and F-wt proteins inhibited cell-cell fusion completely, and no activity was detected. These quantified values were consistent with the levels of syncytium formation analyzed by microscopy (Fig. 2B).
Using this quantitative fusion assay, the effects of truncations of the cytoplasmic tails of the H and F proteins on cell-cell fusion were examined (Fig. 5A). In the absence of the M protein, truncation of the cytoplasmic tail of the F protein increased cell-cell fusion to ∼160% [H-wt, FΔ30, M(−)], indicating that the cytoplasmic tail of the F protein has an inhibitory role in cell-cell fusion, as reported previously (5). The additional expression of the M-wt protein enhanced this fusion even more (H-wt, FΔ30, M-wt), whereas expression of the M-P64S/E89K protein inhibited fusion (H-wt, FΔ30, M-P64S/E89K). Truncation of the cytoplasmic tail of the H protein also increased cell-cell fusion, to ∼192%, in the absence of the M protein [HΔ20, F-wt, M(−)]. However, in contrast to the data for the truncated F protein, neither the M-wt nor the M-P64S/E89K protein modulated the cell-cell fusion activity mediated by the HΔ20 and F-wt proteins. The cell-cell fusion activities in cells expressing HΔ20 and FΔ30 proteins were similar to those in cells expressing HΔ20 and F-wt proteins, regardless of the presence or absence of the M-wt or M-P64S/E89K protein. These data indicate that the presence of the cytoplasmic tails of the H and F proteins of the virulent IC-B strain reduces cell-cell fusion, as observed for the Edmonston vaccine strain (5), and that the M-wt protein enhances and the M-P64S/E89K protein inhibits cell-cell fusion by interacting with the cytoplasmic tail of the H protein of the IC-B strain.
To analyze the effects of truncations of the cytoplasmic tails of the H and F proteins on cell-cell fusion in the context of virus infection, wt and P64S/E89K viruses expressing either the HΔ20 or FΔ30 protein were generated. All viruses were successfully recovered. The wt viruses expressing the HΔ20 or FΔ30 protein produced large syncytia in B95a cells, similar to wt virus (Fig. 5B, top panel). Although the P64S/E89K virus failed to produce syncytia in B95a cells, the P64S/E89K virus expressing the HΔ20 protein produced syncytia that were as large as those produced by the wt virus (Fig. 5B, bottom panel). The P64S/E89K virus expressing the FΔ30 protein also produced syncytia, although they were much smaller than those produced by the wt virus. These data indicate that in the context of virus infection, the M-P64S/E89K protein also inhibits syncytium formation by interacting with the cytoplasmic tail of the H protein.
Roles of the cytoplasmic tails of the H and F proteins in virus growth.
The growth of wt and P64S/E89K viruses expressing a truncated envelope protein (HΔ20 or FΔ30) was examined. Either truncation attenuated the growth of wt virus in B95a cells (Fig. 6A). Truncation of the cytoplasmic tail of the H protein (HΔ20) severely retarded viral growth, but the effect of truncation of the cytoplasmic tail of the F protein (FΔ30) was smaller (Fig. 6A). Since the HΔ20 and FΔ30 proteins exhibited no defect in cell-cell fusion but, rather, showed increased fusion activity in B95a cells (Fig. 5A), the growth attenuation of wt virus expressing the HΔ20 or FΔ30 protein in B95a cells was likely to have been caused by a defect in virus assembly. The cytoplasmic tail of the H protein is likely to be more important than that of the F protein in MV assembly. In contrast, truncation of the cytoplasmic tail of either the H or F protein enhanced MV growth in B95a cells when the recombinant virus also expressed the M-P64S/E89K protein (Fig. 6B). As shown in Fig. 5B, the P64S/E89K virus hardly produced any syncytia in B95a cells, whereas the P64S/E89K virus expressing the HΔ20 or FΔ30 protein produced syncytia. These data suggest that the recovery of cell-cell fusion by truncations in the cytoplasmic tails of the glycoproteins worked advantageously for the P64S/E89K virus, allowing it to spread in B95a cells. Although the growth defect of the P64S/E89K virus in B95a cells was relieved by its possessing the HΔ20 or FΔ30 protein, this virus lost the ability to replicate well in Vero cells (Fig. 6C). Taken together, these data indicate that interactions of the M protein with the cytoplasmic tails of the H and F proteins control virus assembly and cell-cell fusion, modulating MV growth in different cell types.
FIG. 6.
Roles of cytoplasmic tails of the H and F proteins in virus growth. (A) Replication kinetics of recombinant MVs expressing the M-wt protein in B95a cells. Viruses expressed the H or F protein with a truncation in the cytoplasmic domain (HΔ20 or FΔ30). B95a cells were infected with recombinant MVs at an MOI of 0.01, and infectious titers in culture medium (cell-free) and cells (cell-associated) were determined at various intervals. (B) Replication kinetics of recombinant MVs expressing the M-P64S/E89K protein in B95a cells. (C) Replication kinetics of recombinant MVs expressing the M-P64S/E89K protein in Vero cells. Vero cells were infected with recombinant MVs at an MOI of 0.08.
DISCUSSION
MV enters cells efficiently via SLAM and causes extensive syncytium formation as a result of SLAM-dependent membrane fusion between infected cells and neighboring cells. However, inefficient MV entry (but not cell-cell fusion) also occurs in the absence of SLAM (2, 13). Its significance in MV pathogenicity is unclear, but this SLAM-independent entry is important for understanding the mechanisms by which vaccine and laboratory strains of MV adapt to grow in SLAM− cell types, as substitutions in the receptor-binding H protein are not necessarily required for their adaptation (22, 46, 50). We recently showed that the M and L proteins of the Edmonston vaccine strain can confer on SLAM-using MV strains the ability to grow well in Vero cells (45). Among those found in the M protein of the Edmonston strain, two substitutions (P64S and E89K) were identified that were responsible for the efficient growth in Vero cells (45).
In this report, we focused on these P64S and E89K substitutions in the M protein. Indirect immunofluorescence and confocal microscopy revealed that in the absence of other viral components, the M-P64S/E89K protein mostly colocalized with the H protein, forming filamentous structures on the cell surface, whereas the M-wt and H proteins were hardly colocalized. In contrast, both M-wt and M-P64S/E89K proteins partly colocalized with the F protein. These data suggest that the M protein gained the ability to interact with the H protein via the P64S and E89K substitutions, whereas it intrinsically had the ability to interact with the F protein. The P64S and E89K substitutions did not appear to affect the M-F interaction.
Many studies of paramyxoviruses (1, 5, 11, 25, 40, 41, 43, 55) and other negative-strand RNA viruses (41, 62) have reported interactions between the cytoplasmic tails of viral envelope proteins and matrix proteins (although they have not provided direct biochemical evidence of them by using coimmunoprecipitation). Thus, it is presumable that the M-P64S/E89K protein interacts with the cytoplasmic tail of the H protein. When most of the cytoplasmic tail of the H protein was deleted, the M-P64S/E89K protein hardly colocalized with the H protein and failed to form filamentous structures, implying that the M-P64S/E89K protein does indeed interact with the cytoplasmic tail of the H protein. Our attempt to show the direct interaction between M and H proteins by using coimmunoprecipitation was unsuccessful, as were other similar studies (1, 23), suggesting that the interaction is very weak and/or indirect.
It is also suggested that the cytoplasmic tails of envelope proteins or their interactions with M proteins are important for the efficient assembly of paramyxoviruses (6, 8, 9, 40, 41, 51, 52, 55), rhabdoviruses (41, 56), orthomyxoviruses (18, 24, 41), and retroviruses (17, 34). Interestingly, using the Edmonston vaccine strain, Naim et al. showed that the MV M protein controls the sorting of the H and F proteins (26). Therefore, the interaction of the M-P64S/E89K protein with the cytoplasmic tail of the H protein may promote the coalescence of MV proteins at the site of virus budding. Indeed, the P64S/E89K virus produced infectious particles much more efficiently than the wt virus did, although both viruses synthesized similar amounts of viral proteins. Efficient assembly of infectious particles likely contributes to better MV growth in Vero cells. Consistently, the ability of the P64S/E89K virus to grow well in Vero cells was cancelled when the H protein had a truncated cytoplasmic tail.
To have an M protein that efficiently promotes virus assembly seems to be advantageous for MV to grow in cultured cells as well as in vivo. If this is so, then why do clinical isolates of MV have an M protein that supports MV assembly inefficiently? A clue would be the effect on cell-cell fusion. Although the M-P64S/E89K protein was efficient at promoting virus assembly, it inhibited cell-cell fusion. Furthermore, the P64S/E89K virus did not produce large syncytia and replicated less efficiently in B95a cells than the wt virus did. The inhibitory effect of the M-P64S/E89K protein on cell-cell fusion may also operate on virus-cell fusion. However, the P64S/E89K virus replicated well in Vero cells, in which multiple rounds of new infection are critical for efficient growth. These results may suggest a mechanistic difference between virus-cell and cell-cell fusion processes, as suggested for other paramyxoviruses (14, 54). Using Sendai virus, a member of the paramyxovirus family, Henis et al. (14) reported that the lateral mobility of both the F and receptor-binding HN proteins is essential for cell-cell fusion but much less important for virus-cell fusion.
Interestingly, Plemper et al. (35) suggested that the strength of the H and F protein interaction is an important modulator of MV-induced cell-cell fusion. Cathomen et al. (4, 5) reported that recombinant MVs that lacked the M protein or those that expressed H or F proteins with truncated cytoplasmic tails showed increases in cell-cell fusion activity. Thus, the inhibition of cell-cell fusion observed in our study may be caused by the capture of the cytoplasmic tail of the H protein by the M-P64S/E89K protein, which also interacts with the cytoplasmic tail of the F protein. Consistently, when the H protein had a truncated cytoplasmic tail, cell-cell fusion was no longer inhibited by the M-P64S/E89K protein. Furthermore, P64S/E89K virus expressing an H protein with a truncated cytoplasmic tail grew more efficiently in B95a cells than did that expressing the intact H protein. In vivo, MV spreads mainly in the organs of the immune system containing SLAM+ cells (12). Therefore, viruses with the P64S and E89K substitutions in the M protein, which are disadvantageous for MV to spread in SLAM+ cells, are likely to have been eliminated.
MV vaccine strains are obtained by passaging SLAM-using MVs in a variety of cell types (38). One strategy that viruses have taken to adapt to cultured cells is gaining the ability to use CD46 as an alternative receptor (59). However, our studies have indicated that gaining the ability to use CD46 as an efficient receptor is not so easy, because multiple substitutions are usually required (44). In addition, MV vaccine strains were passaged in chicken embryo fibroblasts and sheep kidney and dog kidney cells (38), in which neither SLAM nor CD46 is expressed. Therefore, the original isolates would have had to spread many times in some way via SLAM- and CD46-independent mechanisms. Available sequence data indicate that all MV vaccine strains have both or either of the P64S and E89K substitutions (32, 38, 39) (GenBank accession number EF033071). We speculate that at least one of these substitutions is needed for MV to survive in unnatural host cells.
It should be noted that unlike the P64S/E89K virus, the Edmonston tag strain grows well in B95a cells, producing large syncytia, although it has both P64S and E89K substitutions. The Edmonston tag strain has two other substitutions in the M protein (R175G and A209T) in addition to the P64S and E89K substitutions (45). We found that when the M protein had the R175G substitution, the inhibitory effect on cell-cell fusion due to the P64S and E89K substitutions was partially cancelled (data not shown). The Edmonston tag strain is a recombinant virus generated from cloned cDNAs, and MV strains used to construct the Edmonston tag cDNAs were passaged in CD46+ cell lines (3, 36). Furthermore, the H protein of the Edmonston tag strain can use CD46 efficiently as a receptor (44). From the available sequence data and our biological data, we speculate that the Edmonston tag strain acquired the R175G substitution to restore cell-cell fusion activity when the virus became fully capable of using CD46 as a receptor.
In conclusion, we have demonstrated the roles of the P64S and E89K substitutions found in the M proteins of MV vaccine strains. They promote MV assembly and growth in SLAM− nonlymphoid cells, whereas their inhibitory effect on cell-cell fusion retards MV growth in SLAM+ lymphoid cells. These results have advanced our understanding of the molecular basis for MV adaptation to grow in unnatural host cells and have suggested novel mechanisms by which the interactions between viral matrix proteins and the cytoplasmic tails of envelope glycoproteins actively regulate virus assembly and cell-cell fusion to optimize the pathway of virus spread.
Acknowledgments
We thank E. A. Berger for providing pG1NT7βgal, B. Moss and M. Kohara for recombinant vaccinia viruses, and T. A. Sato, T. Kohama, and R. Cattaneo for providing the antibodies against MV proteins. We are grateful to A. Takade for electron microscopy.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor, and Welfare of Japan and from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Ali, A., and D. P. Nayak. 2000. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology 276:289-303. [DOI] [PubMed] [Google Scholar]
- 2.Andres, O., K. Obojes, K. S. Kim, V. ter Meulen, and J. Schneider-Schaulies. 2003. CD46- and CD150-independent endothelial cell infection with wild-type measles viruses. J. Gen. Virol. 84:1189-1197. [DOI] [PubMed] [Google Scholar]
- 3.Ballart, I., D. Eschle, R. Cattaneo, A. Schmid, M. Metzler, J. Chan, S. Pifko-Hirst, S. A. Udem, and M. A. Billeter. 1990. Infectious measles virus from cloned cDNA. EMBO J. 9:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattaneo, R., and J. K. Rose. 1993. Cell fusion by the envelope glycoproteins of persistent measles viruses which caused lethal human brain disease. J. Virol. 67:1493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coronel, E. C., T. Takimoto, K. G. Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolganiuc, V., L. McGinnes, E. J. Luna, and T. G. Morrison. 2003. Role of the cytoplasmic domain of the Newcastle disease virus fusion protein in association with lipid rafts. J. Virol. 77:12968-12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouillot-Coriou, N., and L. Roux. 2000. Structure-function analysis of the Sendai virus F and HN cytoplasmic domain: different role for the two proteins in the production of virus particle. Virology 270:464-475. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghildyal, R., D. Li, I. Peroulis, B. Shields, P. G. Bardin, M. N. Teng, P. L. Collins, J. Meanger, and J. Mills. 2005. Interaction between the respiratory syncytial virus G glycoprotein cytoplasmic domain and the matrix protein. J. Gen. Virol. 86:1879-1884. [DOI] [PubMed] [Google Scholar]
- 12.Griffin, D. E. 2001. Measles virus, p. 1401-1441. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 13.Hashimoto, K., N. Ono, H. Tatsuo, H. Minagawa, M. Takeda, K. Takeuchi, and Y. Yanagi. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henis, Y. I., Y. Herman-Barhom, B. Aroeti, and O. Gutman. 1989. Lateral mobility of both envelope proteins (F and HN) of Sendai virus in the cell membrane is essential for cell-cell fusion. J. Biol. Chem. 264:17119-17125. [PubMed] [Google Scholar]
- 15.Hirano, A., M. Ayata, A. H. Wang, and T. C. Wong. 1993. Functional analysis of matrix proteins expressed from cloned genes of measles virus variants that cause subacute sclerosing panencephalitis reveals a common defect in nucleocapsid binding. J. Virol. 67:1848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber, M., R. Cattaneo, P. Spielhofer, C. Orvell, E. Norrby, M. Messerli, J. C. Perriard, and M. A. Billeter. 1991. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299-308. [DOI] [PubMed] [Google Scholar]
- 17.Januszeski, M. M., P. M. Cannon, D. Chen, Y. Rozenberg, and W. F. Anderson. 1997. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 71:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kashiwakuma, T., A. Hasegawa, T. Kajita, A. Takata, H. Mori, Y. Ohta, E. Tanaka, K. Kiyosawa, T. Tanaka, S. Tanaka, N. Hattori, and M. Kohara. 1996. Detection of hepatitis C virus specific core protein in serum of patients by a sensitive fluorescence enzyme immunoassay (FEIA). J. Immunol. Methods 190:79-89. [DOI] [PubMed] [Google Scholar]
- 20.Kobune, F., H. Sakata, and A. Sugiura. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobune, F., H. Takahashi, K. Terao, T. Ohkawa, Y. Ami, Y. Suzaki, N. Nagata, H. Sakata, K. Yamanouchi, and C. Kai. 1996. Nonhuman primate models of measles. Lab. Anim. Sci. 46:315-320. [PubMed] [Google Scholar]
- 22.Kouomou, D. W., and T. F. Wild. 2002. Adaptation of wild-type measles virus to tissue culture. J. Virol. 76:1505-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markwell, M. A., and C. F. Fox. 1980. Protein-protein interactions within paramyxoviruses identified by native disulfide bonding or reversible chemical cross-linking. J. Virol. 33:152-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitnaul, L. J., M. R. Castrucci, K. G. Murti, and Y. Kawaoka. 1996. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 70:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moll, M., H. D. Klenk, and A. Maisner. 2002. Importance of the cytoplasmic tails of the measles virus glycoproteins for fusogenic activity and the generation of recombinant measles viruses. J. Virol. 76:7174-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naim, H. Y., E. Ehler, and M. A. Billeter. 2000. Measles virus matrix protein specifies apical virus release and glycoprotein sorting in epithelial cells. EMBO J. 19:3576-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakatsu, Y., M. Takeda, M. Kidokoro, M. Kohara, and Y. Yanagi. 2006. Rescue system for measles virus from cloned cDNA driven by vaccinia virus Lister vaccine strain. J. Virol. Methods 137:152-155. [DOI] [PubMed] [Google Scholar]
- 28.Nakatsu, Y., M. Takeda, S. Ohno, R. Koga, and Y. Yanagi. 2006. Translational inhibition and increased interferon induction in cells infected with a C protein-deficient measles virus. J. Virol. 80:11861-11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 30.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono, N., H. Tatsuo, Y. Hidaka, T. Aoki, H. Minagawa, and Y. Yanagi. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks, C. L., R. A. Lerch, P. Walpita, H. P. Wang, M. S. Sidhu, and S. A. Udem. 2001. Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75:910-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. Davidson and R. M. Elliott (ed.), Molecular virology: a practical approach. IRL Oxford University Press, Oxford, United Kingdom.
- 34.Piller, S. C., J. W. Dubay, C. A. Derdeyn, and E. Hunter. 2000. Mutational analysis of conserved domains within the cytoplasmic tail of gp41 from human immunodeficiency virus type 1: effects on glycoprotein incorporation and infectivity. J. Virol. 74:11717-11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson, C. D., A. Scheid, and P. W. Choppin. 1980. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology 105:205-222. [DOI] [PubMed] [Google Scholar]
- 38.Rota, J. S., Z. D. Wang, P. A. Rota, and W. J. Bellini. 1994. Comparison of sequences of the H, F, and N coding genes of measles virus vaccine strains. Virus Res. 31:317-330. [DOI] [PubMed] [Google Scholar]
- 39.Santos, P. R., M. L. Azevedo, M. B. Borges, M. S. Freire, J. P. Nascimento, and M. T. Moraes. 2003. Comparative sequence analysis of the P-, M- and L-coding region of the measles virus CAM-70 live attenuated vaccine strain. Braz. J. Med. Biol. Res. 36:1475-1484. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt, A. P., B. He, and R. A. Lamb. 1999. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J. Virol. 73:8703-8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt, A. P., G. P. Leser, D. L. Waning, and R. A. Lamb. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spielhofer, P., T. Bachi, T. Fehr, G. Christiansen, R. Cattaneo, K. Kaelin, M. A. Billeter, and H. Y. Naim. 1998. Chimeric measles viruses with a foreign envelope. J. Virol. 72:2150-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tahara, M., M. Takeda, F. Seki, T. Hashiguchi, and Y. Yanagi. 2007. Multiple amino acid substitutions in hemagglutinin are necessary for wild-type measles virus to acquire the ability to use receptor CD46 efficiently. J. Virol. 81:2564-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahara, M., M. Takeda, and Y. Yanagi. 2005. Contributions of matrix and large protein genes of the measles virus Edmonston strain to growth in cultured cells as revealed by recombinant viruses. J. Virol. 79:15218-15225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeda, M., A. Kato, F. Kobune, H. Sakata, Y. Li, T. Shioda, Y. Sakai, M. Asakawa, and Y. Nagai. 1998. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J. Virol. 72:8690-8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeda, M., S. Ohno, F. Seki, K. Hashimoto, N. Miyajima, K. Takeuchi, and Y. Yanagi. 2005. Efficient rescue of measles virus from cloned cDNA using SLAM-expressing Chinese hamster ovary cells. Virus Res. 108:161-165. [DOI] [PubMed] [Google Scholar]
- 48.Takeda, M., S. Ohno, F. Seki, Y. Nakatsu, M. Tahara, and Y. Yanagi. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J. Virol. 79:14346-14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeda, M., K. Takeuchi, N. Miyajima, F. Kobune, Y. Ami, N. Nagata, Y. Suzaki, Y. Nagai, and M. Tashiro. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74:6643-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi, K., N. Miyajima, F. Kobune, and M. Tashiro. 2000. Comparative nucleotide sequence analysis of the entire genomes of B95a cell-isolated and Vero cell-isolated measles viruses from the same patient. Virus Genes 20:253-257. [DOI] [PubMed] [Google Scholar]
- 51.Takimoto, T., T. Bousse, E. C. Coronel, R. A. Scroggs, and A. Portner. 1998. Cytoplasmic domain of Sendai virus HN protein contains a specific sequence required for its incorporation into virions. J. Virol. 72:9747-9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao, T., M. H. Skiadopoulos, F. Davoodi, J. M. Riggs, P. L. Collins, and B. R. Murphy. 2000. Replacement of the ectodomains of the hemagglutinin-neuraminidase and fusion glycoproteins of recombinant parainfluenza virus type 3 (PIV3) with their counterparts from PIV2 yields attenuated PIV2 vaccine candidates. J. Virol. 74:6448-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tatsuo, H., N. Ono, K. Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893-897. [DOI] [PubMed] [Google Scholar]
- 54.Waning, D. L., C. J. Russell, T. S. Jardetzky, and R. A. Lamb. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. USA 101:9217-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waning, D. L., A. P. Schmitt, G. P. Leser, and R. A. Lamb. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 76:9284-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitt, M. A., L. Chong, and J. K. Rose. 1989. Glycoprotein cytoplasmic domain sequences required for rescue of a vesicular stomatitis virus glycoprotein mutant. J. Virol. 63:3569-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wild, T. F., E. Malvoisin, and R. Buckland. 1991. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J. Gen. Virol. 72:439-442. [DOI] [PubMed] [Google Scholar]
- 58.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]
- 59.Yanagi, Y., M. Takeda, and S. Ohno. 2006. Measles virus: cellular receptors, tropism and pathogenesis. J. Gen. Virol. 87:2767-2779. [DOI] [PubMed] [Google Scholar]
- 60.Yanagi, Y., M. Takeda, S. Ohno, and F. Seki. 2006. Measles virus receptors and tropism. Jpn. J. Infect. Dis. 59:1-5. [PubMed] [Google Scholar]
- 61.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]