Abstract
Apoptotic cell death forms part of the host defense against virus infection. We tested orf virus, a member of the poxvirus family, for the ability to inhibit apoptosis and found that orf virus-infected cells were fully resistant to UV-induced changes in cell morphology, caspase activation, and DNA fragmentation. By using a library of vaccinia virus-orf virus recombinants, we identified an orf virus gene (ORFV125) whose presence was linked with the inhibition of apoptosis. The 173-amino-acid predicted protein had no clear homologs in public databases other than those encoded by other parapoxviruses. However, ORFV125 possessed a distinctive C-terminal domain which was necessary and sufficient to direct the protein to the mitochondria. We determined that ORFV125 alone could fully inhibit UV-induced DNA fragmentation, caspase activation, and cytochrome c release and that its mitochondrial localization was required for its antiapoptotic function. In contrast, ORFV125 did not prevent UV-induced activation of c-Jun NH2-terminal kinase, an event occurring upstream of the mitochondria. These features are comparable to the antiapoptotic properties of the mitochondrial regulator Bcl-2. Furthermore, bioinformatic analyses revealed sequence and secondary-structure similarities to Bcl-2 family members, including characteristic residues of all four Bcl-2 homology domains. Consistent with this, the viral protein inhibited the UV-induced activation of the proapoptotic Bcl-2 family members Bax and Bak. ORFV125 is the first parapoxvirus apoptosis inhibitor to be identified, and we propose that it is a new antiapoptotic member of the Bcl-2 family.
Apoptosis is a vital process within multicellular organisms which removes cells that are old, unwanted, or potentially dangerous. It plays pivotal roles in the development and homeostasis of tissues, as well as in immune responses to pathological signals, including virus infection (25, 28). Two major apoptotic pathways have been identified that involve either the binding of “death ligands” to cell surface receptors (extrinsic pathway) or the activation of mitochondria and endoplasmic reticulum (ER) by various stress signals (intrinsic pathway). Both pathways result in the activation of caspases which subsequently cleave a large number of cellular proteins, leading to cell death.
The mitochondrial pathway is characterized by the permeabilization of the outer mitochondrial membrane and the release of proapoptotic factors such as cytochrome c from the intermembrane space into the cytoplasm. The mechanism by which these proapoptotic factors are released is still controversial (8, 15, 16). Mitochondrial integrity is regulated by members of the Bcl-2 family of proteins (3, 53). Proapoptotic Bcl-2 family members most likely interact with the mitochondrial membrane to induce the release of proteins into the cytoplasm, whereas antiapoptotic members inhibit the activity of their proapoptotic counterparts.
The Bcl-2 family members are α-helical proteins with seven to nine helices, and their defining characteristic is the presence of at least one of four regions of sequence homology referred to as Bcl-2 homology (BH) domains. While antiapoptotic members such as Bcl-2, Bcl-xL, or Bcl-w have all four domains, proapoptotic proteins share either the first three BH domains (Bax, Bak, and Bok) or only the BH3 domain (e.g., Bad, Bim, Bik, or Bid). A prominent hydrophobic groove, which is formed by the BH1, BH2, and BH3 domains, is present on the surface of antiapoptotic and Bax-like proapoptotic Bcl-2 proteins and serves as a binding site for the α-helical BH3 domain of proapoptotic family members (39). Structural analysis and mutagenesis studies of conserved residues within the BH domains and the hydrophobic groove reveal that these domains are important not only for the formation of hetero- and homocomplexes but also for the pro- or antiapoptotic function of Bcl-2 family members (17, 39). In addition to the BH domains, most family members exhibit a C-terminal targeting motif which is responsible for directing the proteins to intracellular membranes, including the mitochondrial, nuclear, and ER membranes (44).
Apoptosis plays important roles in host defenses against virus infection. Infected cells may recognize virus particles at cell entry, viral proteins and DNA/RNA during early viral replication, or virus-induced alterations to cellular physiology and, in response, execute the cell suicide program in order to block virus replication (11). It is therefore not surprising that viruses have developed a vast array of modulators that block apoptosis at different stages within the apoptotic pathways, for instance, caspase inhibitors, environmental stress response inhibitors, proteins that interfere with the death receptor pathway or the interferon pathway, and mitochondrial modulators (11, 12). The latter group includes viral Bcl-2 homologs, which have been identified in adenoviruses, herpesviruses, and poxviruses. The sequence identity of cellular and viral Bcl-2 proteins is low, but the secondary and tertiary structures are conserved. Viral Bcl-2 proteins are α-helical proteins with a conserved BH1 domain and at least one other BH domain, either a BH2 or a BH3 domain, while most of them lack the BH4 domain (6, 20). Two viral Bcl-2 homologs have been described within the poxvirus family, FWPV039 from fowlpox virus and its ortholog CNPV058 from canarypox virus (1, 49). However, a functional analysis of these genes has not been reported. The poxvirus family encodes additional mitochondrion-localized antiapoptotic proteins which have not been regarded as Bcl-2 family members, M11L from myxoma virus (10) and F1L from vaccinia virus (VACV) (52).
Orf virus (ORFV) is a member of the poxvirus family belonging to the Parapoxvirus genus. It causes a highly contagious, eruptive skin disease in sheep and goat populations, can be transmitted to humans, and is able to reinfect hosts that have been previously infected (14). Its 140-kb linear, double-stranded DNA genome consists of a central core which is highly conserved among vertebrate poxviruses and includes information essential for replication within the host cell and more variable outer regions that encode functions often not required for replication in vitro but which play important roles during infection in vivo. Interestingly, the outer regions of the ORFV genome differ substantially from those of other poxviruses. The 25-kb right terminal region of the ORFV genome contains a number of unique parapoxvirus genes, including an interleukin-10 gene, a granulocyte-macrophage colony-stimulating factor binding protein gene, a chemokine-binding protein gene, and a vascular endothelial growth factor gene (14, 34). In addition, ORFV and the other parapoxviruses lack obvious counterparts of the antiapoptotic factors seen in other poxviruses, apart from a homolog of VACV E3L (33), a double-stranded-RNA-binding protein present in many poxviruses.
In this report, we show that ORFV expresses antiapoptotic activity and identify the gene responsible (ORFV125). The encoded protein localizes to and functions at the mitochondria to fully block downstream apoptotic events. Furthermore, it not only acts in a similar manner but also shares key sequence elements with members of the Bcl-2 family, suggesting that ORFV125 is a Bcl-2-like protein.
MATERIALS AND METHODS
Cells.
143B cells and derivatives were grown in Eagle minimal essential medium (Sigma catalog no. M0769), while HeLa cells were cultured in Dulbecco's modified Eagle medium (Sigma catalog no. 058). All media were supplemented with 10% fetal bovine serum (GIBCO), 2 mM l-glutamine, 500 U/ml penicillin, 500 μg/ml streptomycin, and 100 μg/ml kanamycin (Roche). For construction of stable cell lines expressing Bcl-2, ORFV125, or the empty vector, 2 × 106 143B cells were transfected with 4 μg of plasmid DNA (pEF_FLAG-Bcl-2PGKpuro, pEF_FLAG-ORFV125PGKpuro, or pEF_FLAGPGKpuro) by using Lipofectin (Invitrogen Life Technologies) according to the manufacturer's protocol. To select for cells carrying the constructs, 0.5 μg/ml puromycin was added 24 h after transfection. Single clones were isolated by limiting dilution, and their identities were confirmed by sequence analysis of the inserted DNA and expression of the FLAG-tagged proteins.
Wild-type and recombinant viruses.
ORFV strain NZ2 (41), VACV strain Lister (42), and VACV strain MVA (2) were used in this study. The construction and characterization of a library of VACV-ORFV recombinants (VVOV216, -80, -213, -97, -96, -212, -245, -86, -243, -283, -330, -85, and -82; see Fig. 2A) were previously described (35). In this study, three additional recombinants (VVOV214, -630, and -629; see Fig. 2C) were constructed in the same manner (35), with plasmids pVU214, pVU630, and pVU629, respectively.
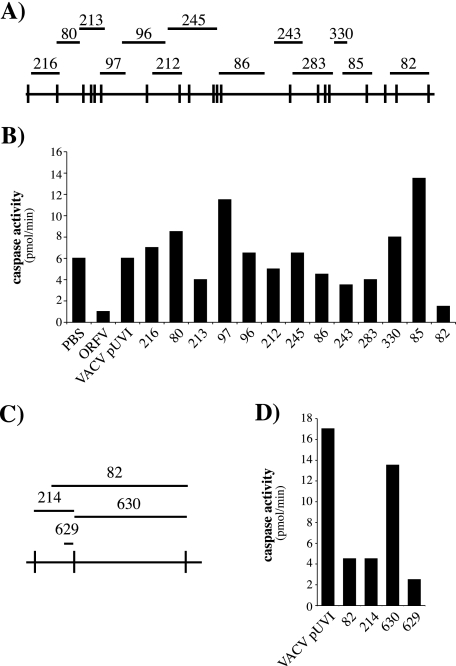
FIG. 2.
Use of VACV-ORFV recombinants to map an ORFV inhibitor of UV-induced apoptosis. (A) The ORFV KpnI map is shown at the bottom, and the bars above indicate the locations of DNA fragments that were recombined into the VACV genome to create the VVOV recombinant library. (B) 143B cells were mock infected (PBS) or infected, at a multiplicity of infection of 4, with ORFV or the indicated VACV recombinants and treated with UV 7 h later. After 12 h, the samples were analyzed for caspase activity. The VACV recombinant pUVI is a control virus carrying no ORFV DNA (35). (C) An expanded view of the right end of the ORFV KpnI map with lines above showing the relative locations of the DNA fragments used to generate VVOV82, -214, 630, and -629. (D) Caspase activities in UV-irradiated 143B cells infected with the indicated VACV recombinants and prepared as described for panel B.
Plasmid construction. (i) Plasmids for creating VACV-ORFV recombinants.
For the construction of pVU214, the KpnI K fragment (35) of the ORFV genome was inserted into the transfer vector pUVI (13). pVU630 was created by cloning the ORFV KpnI E fragment (35) into pVU433, a derivate of pUVI from which the P11 transcriptional promoter sequence has been removed. A linker created by annealing two partially complementary oligonucleotides, 5′EcoRI-AATTCGAACCCCTTCG and 5′HindIII-AGCTCGAAGGGGTTCG, was ligated with EcoRI-digested pUVI, and the mixture was then digested with HindIII. The two larger products (3.7 and 4.3 kb) were purified and ligated. A clone in which the relative orientation of these two fragments was the same as in pUVI was recovered and named pVU433. pVU629 was generated by inserting a 729-bp PCR product containing the ORFV125 open reading frame and its own promoter region into the EcoRI and BamHI sites of pVU433. The PCR product was amplified from ORFV NZ2 DNA with primers 5′EcoRI-CGAATTCCGAGGAAGAAGAGGAGGAA and 5′BamHI-CGGGATCCAATGTCGTATTTCAGTTTGA.
(ii) GFP fusion constructs.
The ORFV125 coding region was amplified from ORFV NZ2 DNA with primers 5′BglII-CGAGATCTATGGCAAACAGAGAAGAG and 5′BamHI-TAGGATCCTTATGTGCGCCGCAACAC. The PCR product was cleaved with BglII/BamHI and cloned into BglII/BamHI-digested phrGFPN1(Stratagene) to create phrGFPN1-ORFV125wt. With this plasmid as a PCR template, we generated phrGFPN1-ORFV125Δts (primers 5′BglII-CGAGATCTATGGCAAACAGAGAAGAG and 5′HindIII-CGAAGCTTTTACACGCGCGCGACGGC) and phrGFPN1-ORFV125ts (primers 5′BglII-ATAGATCTCGGCGCCGCCTGCACC and 5′HindIII-CGAAGCTTTTATGTGCGCCGCAACAC). To achieve brighter green fluorescent protein (GFP) fluorescence, these three open reading frames were then subcloned into pEGFP-C1 (BD Bioscience Clontech) with BspEI and BamHI to create pEGFPC1-ORFV125wt, pEGFPC1-ORFV125Δts, and pEGFPC1-ORFV125ts.
For construction of phrGFPN1-Bcl-w, the Bcl-w coding region was amplified from pGEX-6P-3-Bcl-w with primers 5′BglII-GCCAGATCTATGGCGACCCCAGCCTCG and 5′EcoRI-GGGAATTCACTTGCTAGCAAAAAAGGC and cloned into phrGFPN1 to create a GFP-Bcl-w fusion protein. The Bcl-w ORF was subcloned into pEGFP-C1 with BspEI and EcoRI to create pEGFPC1-Bcl-w. The Bcl-2 coding region was amplified from pEF_Bcl-2PGKpuro (23) with 5′BspEI-GGTCCGGAATGGCGCACGCTGGGAGAACAG and 5′EcoRI-CCGAATTCTCACTTGTGGCCCAGATAGGC and directly cloned into pEGFP-C1 to create pEGFPC1-Bcl-2.
(iii) Plasmids for creating stable cell lines.
The three constructs pEF_FLAG-ORFV125PGKpuro, pEF_FLAG-Bcl-2PGKpuro, and pEF_FLAGPGKpuro were produced by replacing the resident Bcl-2 gene in pEF_Bcl-2PGKpuro (23) with FLAG-tagged cDNAs of ORFV125 and Bcl-2 or a FLAG element, respectively. The FLAG-tagged Bcl-2 coding region was inserted as a PCR product which had been amplified from pEF_Bcl-2PGKpuro with 5′BglII-GGAGATCTATGGACTACAAGGACGACGATGACAAGATGGCGCACGCTGGGAGAACAG and 5′XbaI-CCTCTAGACTCACTTGTGGCCCAGATAGGC. The FLAG-tagged ORFV125 coding region was amplified from pEGFPC1-ORFV125wt with primers 5′BglII-GGAGATCTATGGACTACAAGGACGACGATGACAAGATGGCAAACAGAGAAGAGATTG and 5′XbaI-CCTCTAGATTATGTGCGCCGCAACACGC and digested with BglII and XbaI. The eight-codon-long FLAG element flanked by BglII and XbaI sites was created from two partially complementary oligonucleotides (5′-GATCCATGGACTACAAGGACGACGATGACAAGTAAT and 5′-CTAGATTACTTGTCATCGTCGTCCTTGTAGTCCATG).
Confocal and fluorescence microscopy.
HeLa cells were grown in six-well plates on coverslips and transfected with 1 μg DNA/106 cells with Lipofectin (Invitrogen Life Technologies) according to the manufacturer's protocol. Twenty-four hours later, the cells were stained with 50 nM Mitotracker Red CMXRos (Molecular Probes) for 30 min at 37°C and analyzed with an LSM510 laser scanning confocal microscope. GFP and Mitotracker fluorescence was excited with wavelengths of 543 and 489 nm, respectively.
For the detection of active Bax or Bak, 143B stable cell lines were fixed with 4% paraformaldehyde, permeabilized with 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and blocked in phosphate-buffered saline (PBS) containing 20% fetal bovine serum. Cells were then incubated with antibodies specific for an N-terminal epitope of either Bax (1:250, 6A7; BD PharMingen catalog no. 556467) or Bak (1:250, TC-100; Calbiochem catalog no. AM03), and bound antibody was detected with anti-mouse Alexa Fluor 488 (1:100; Molecular Probes). After mounting in SlowFade (Molecular Probes), the cells were examined with an Olympus BX51 fluorescence microscope. The anti-Bax and anti-Bak antibodies recognize epitopes that only become exposed when Bax or Bak is activated.
Induction of apoptosis.
Cells were UV irradiated by inverting a drained 35-mm dish onto a transilluminator and exposing the cells for 4 s to 20 W/m2 (80 J/m2) UVC light (260 nm). Medium was added, and the cells were incubated for various times at 37°C.
Detection of DNA fragmentation.
To visualize DNA laddering, UV-irradiated cells were harvested by a protocol described previously (32). Briefly, floating and adherent cells were lysed with 200 μl Cartier's buffer (10 mM EDTA; 10 mM Tris-HCl, pH 7.5; 0.2% Triton X-100) (5) for 20 min at room temperature and extracted once with 1 volume phenol and twice with 1 volume chloroform-isoamyl alcohol. DNA was ethanol precipitated and resuspended in 20 μl H2O (plus 100 μg/ml RNase) before electrophoresis on a 1% agarose gel.
For analyzing the DNA content of cells by flow cytometry, cells were harvested as previously described (9). Briefly, floating and adherent cells were collected and resuspended in fluorescence-activated cell sorter buffer (0.1% glucose in PBS). One million cells were processed for each analysis. GFP-transfected cells were fixed with 1% formaldehyde solution (in PBS) on ice for 1 h and washed twice with ice-cold PBS before permeabilization with 70% ethanol overnight at 4°C. Virus-infected cells and stable cell lines were fixed and permeabilized in 70% ethanol without prior treatment. After fixation, cells were resuspended in staining solution (50 μg/ml propidium iodide [PI] and 100 U/ml RNase in PBS), incubated at room temperature for at least 30 min, and analyzed within 3 h with a Becton Dickinson FACScalibur. After exclusion of doublets and cell aggregates, the PI intensities of 10,000 single PI-positive cells in non-GFP experiments or at least 2,000 single PI- and GFP-positive cells in GFP experiments were displayed in an FL2 area histogram. To ensure that any apparent functional differences between the GFP constructs did not result from differences in expression levels, we restricted our analysis to populations with similar GFP expression levels (geometric mean fluorescence intensities). The percentage of cells located in the sub-G0/G1 peak (hypodiploid cells) was quantified with the CellQuest Pro analysis program from BD Bioscience (version 5.2).
Measurement of caspase activity.
143B cells were tested for caspase activity as described in reference 24. Briefly, 2.5 × 105 cells were washed with PBS and the cell pellet was snap-frozen in liquid nitrogen and quickly thawed in 100 μl 37°C prewarmed buffer containing 100 mM HEPES, 10% (wt/vol) sucrose, 5 mM dithiothreitol, and 0.1% CHAPS (pH 7.25) and supplemented with a 25 μM concentration of the caspase substrate Ac-DEVD-AMC (Calbiochem). The fluorescent product aminomethylcoumarin (AMC) was quantified with a Polarstar optima fluorescent plate reader (BMG Labtechnologies; excitation and emission wavelengths, 390 and 460 nm, respectively) at 37°C. The caspase activity of the total sample, in picomoles per minute, was calculated by using an AMC standard curve.
In vitro caspase activation assay.
Cytosolic extracts (S-100) were prepared by a method similar to that described in reference 31. Briefly, 8 × 107 143B cells/ml, resuspended in buffer containing 20 mM HEPES (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 1 mM dithiothreitol, and a protease inhibitor cocktail (complete, mini, EDTA free; Roche), were lysed by repeated passage through a 26-gauge needle. Nuclei were removed by centrifugation (10 min at 1,000 × g and 4°C). The supernatant was further centrifuged for 1 h at 100,000 × g and 4°C to obtain the cytosolic extract (S-100). The total protein amount was measured with the bicinchoninic acid assay (Pierce). For the assay of caspase activation (19), 150 μg S-100 was incubated in caspase buffer (see above) in the presence of 1 mM dATP, 50 μM Ac-DEVD-AMC, and various concentrations of cytochrome c. The fluorescent product AMC was quantified as described above.
Cytochrome c release assay.
143B cells were fractionated into cytosolic and mitochondrial fractions as described elsewhere (24). To achieve this, the pellet of 5 × 105 cells was resuspended in 30 μl ice-cold STE buffer (0.25 M sucrose, 5 mM Tris, 1 mM EGTA, pH 7.4) containing a protease inhibitor cocktail (complete, mini, EDTA free; Roche) and 7.5 μg digitonin (BDH; 30 μg digitonin/mg protein). After a 10-min incubation step on ice, samples were centrifuged for 10 min at 13,000 × g and 4°C. The supernatant was retained as the cytosolic fraction, while the pellet was treated with 30 μl PBS supplemented with 0.5% Triton X-100 for 5 min on ice. The samples were then centrifuged for 10 min at 13,000 × g at 4°C, and the supernatant, representing the mitochondrial fraction, was retained. One-third of each sample was examined for cytochrome c by immunoblotting with a mouse anti-cytochrome c antibody (clone 7H8.2C12; Pharmingen) and an anti-mouse horseradish peroxidase (HRP)-conjugated antibody (Bio-Rad).
Measurement of c-Jun NH2-terminal kinase (JNK) activation.
143B cells (4 × 105) were seeded in 35-mm dishes. At different time points after UV treatment (see above), cells were lysed in 100 μl sodium dodecyl sulfate sample buffer (62.5 mM Tris-HCl, pH 6.8; 2% sodium dodecyl sulfate; 10% glycerol; 0.01% bromophenol blue; 10% β-mercaptoethanol) at 95°C for 5 min and centrifuged for 1 min. Twenty microliters of each supernatant was analyzed by immunoblotting with rabbit phospho-SAPK-JNK and an anti-rabbit, HRP-conjugated secondary antibody (both from Cell Signaling). Total JNK was detected with a mouse anti-JNK1/JNK2 antibody (BD Pharmingen) and an anti-mouse HRP-conjugated secondary antibody (Bio-Rad).
Bioinformatic analysis.
Protein comparisons were conducted with the BLAST suite of programs (http://www.ncbi.nlm.nih.gov/BLAST/) and the HHpred program (http://toolkit.tuebingen.mpg.de/sections/search#). The TMHMM program was used to predict transmembrane (TM) domains (http://www.cbs.dtu.dk/services/TMHMM/), and the Jpred web server was used for prediction of secondary structures (http://www.compbio.dundee.ac.uk/∼www-jpred/). Multiple sequence alignments were performed with ClustalW (http://www.ebi.ac.uk/clustalw/) and adjusted manually according to known structures.
Statistical analysis.
A one-way analysis of variance with a Tukey-Kramer multiple-comparison test was performed with GraphPad InStat version 3.0a for Macintosh (www.graphpad.com).
RESULTS
ORFV infection inhibits apoptosis.
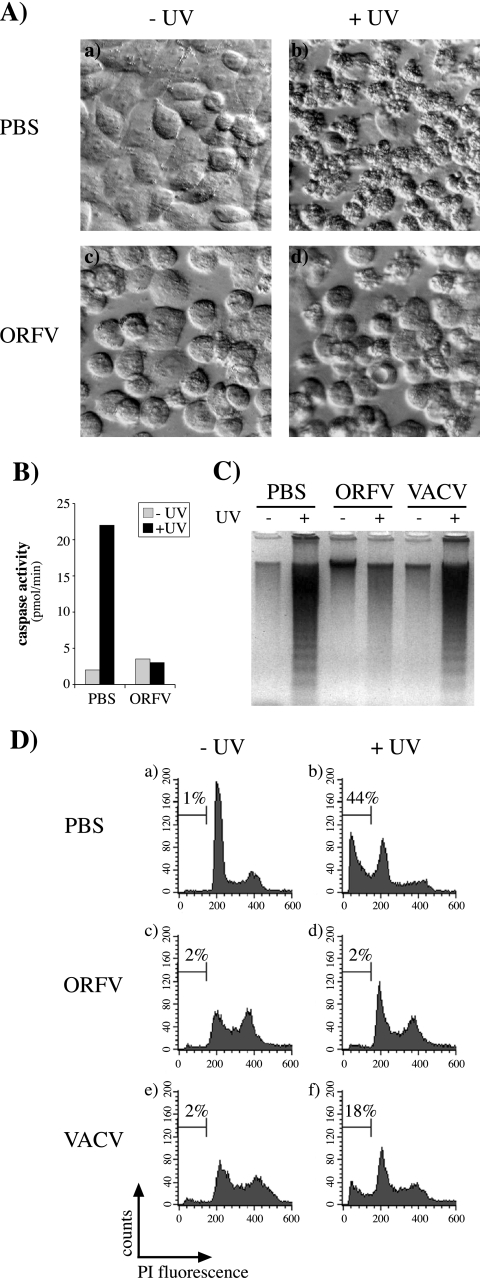
In order to test if ORFV could inhibit the induction of apoptosis, HeLa cells were infected with ORFV, UV irradiated 7 h later, and examined after a further 14 h. Many of the uninfected, UV-irradiated cells showed pronounced membrane blebbing consistent with apoptosis (Fig. 1A). In contrast, ORFV-infected cells displayed cell rounding typical of the cytopathic effects of infection but showed markedly less evidence of an apoptotic response to UV irradiation. Similarly, 143B cells infected with ORFV showed no induction of caspase activity when UV irradiated, whereas a substantial increase in caspase activity was seen in irradiated, uninfected cells (Fig. 1B). The ability of ORFV to inhibit apoptosis was next compared with that of VACV. DNA laddering is a classic indicator of apoptosis and was clearly evident in lysates of 143B cells treated with UV, but cells infected with ORFV were resistant to this apoptotic stimulus (Fig. 1C). In contrast, VACV appeared not to inhibit DNA fragmentation. This was surprising since VACV encodes characterized inhibitors of apoptosis, including one demonstrated to inhibit UV-induced apoptosis (4). To investigate this difference, a more quantitative analysis of the DNA fragmentation response was undertaken by flow cytometry analysis of permeabilized, PI-stained cells. This analysis showed that the number of ORFV-infected cells with a DNA content of less than 2n (apoptotic cells) did not increase in response to UV irradiation (Fig. 1D), whereas with uninfected cells the apoptotic population increased from a background level of 2% to 44%. The response of VACV-infected cells was intermediate (18%), consistent with a significant but partial inhibition of UV-induced apoptosis. ORFV also prevented UV-induced apoptosis in a range of other cells, including primary lamb testis cells, in which ORFV completes productive replication. In addition, ORFV infection significantly inhibited apoptosis induced by staurosporine, actinomycin D, and a combination of tumor necrosis factor and cycloheximide (results not shown).
FIG. 1.
ORFV inhibits UV-induced apoptosis. Cells were mock infected (PBS) or infected with ORFV or VACV, either strain MVA (C) or Lister (D), at a multiplicity of infection of 4 and UV irradiated (+UV) 7 h later. Cells not irradiated are indicated by −UV. (A) At 14 h after UV treatment, HeLa cells were examined with an inverted microscope (Olympus; Zeiss) equipped with (20×) Hoffmann modulation contrast optics. (B) Caspase activity in 143B cell lysates prepared 12 h after UV treatment. (C) DNA laddering was visualized by agarose gel electrophoresis of 143B cell lysates prepared 17 h after UV treatment. (D) At 17 h after UV irradiation, the DNA content of permeabilized, PI-stained 143B cells was analyzed by flow cytometry. The percentage of hypodiploid (apoptotic) cells in each population is indicated on each histogram.
Screening for the ORFV antiapoptotic gene.
Initially we found that UV-inactivated ORFV retained the full inhibitory activity of viable virus, suggesting that the ORFV gene(s) responsible was expressed prior to DNA replication. In order to locate the ORFV gene(s), we took advantage of VACV's partial inhibition of UV-induced apoptosis and screened VACV-ORFV recombinants to identify an ORFV gene(s) that might bestow full inhibition on VACV. Cells were infected with 13 VACV recombinants, each containing a defined fragment of the ORFV genome, and all of them together represented >95% of the genome in overlapping fragments (Fig. 2A) (35). Infected cells were UV irradiated and 12 h later assayed for caspase activity.
The results, representative of three independent experiments, are shown in Fig. 2B. Only 1 of the 13 recombinants, VVOV82, showed the same inhibitory activity as ORFV. The VVOV82 recombinant genome includes a 13.7-kb fragment of ORFV DNA that encodes 10 genes (ORFV124 to ORFV134) (34). We tested VACV recombinants carrying smaller fragments from this particular region (Fig. 2C and D). Recombinant VVOV630, carrying the 10.3-kb KpnI E fragment, had no inhibitory activity, whereas VVOV214, containing the adjacent 3.8-kb KpnI K fragment, was fully inhibitory. We further demonstrated that a VACV recombinant (VVOV629) carrying only a single gene from the KpnI K fragment (ORFV125) had the same inhibitory activities as ORFV, VVOV82, and VVOV214 (Fig. 2C and D).
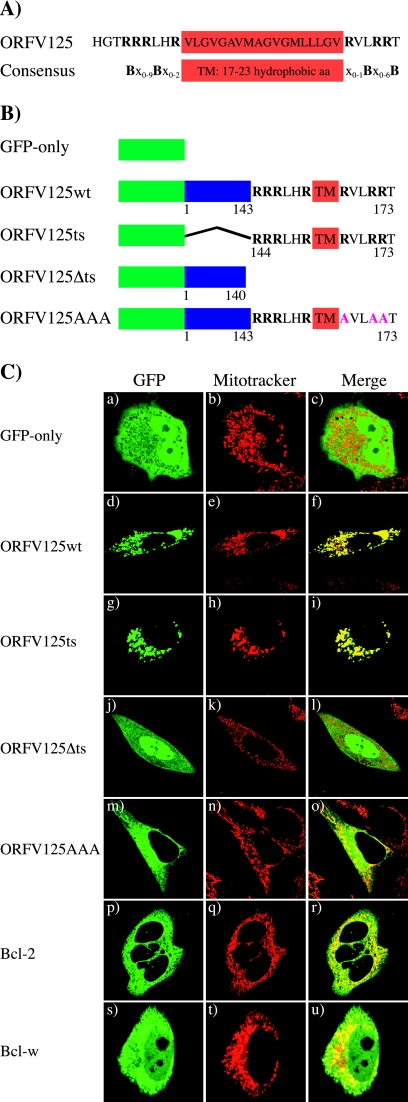
The ORFV protein ORFV125 exhibits a mitochondrion-targeting sequence that directs the protein to the mitochondria.
ORFV125 is flanked by transcriptional motifs typical of poxvirus early genes and is predicted to encode a 173-amino-acid, 19-kDa protein (34). Comparisons of the ORFV125 peptide sequence with public databases did not reveal any homologous proteins other than orthologs from other parapoxviruses. However, sequence analysis revealed a putative mitochondrion-targeting sequence in the C-terminal region, consisting of a predicted TM domain flanked by positively charged amino acids (Fig. 3A). This motif is conserved within tail-anchored proteins such as TOM-5 and TOM-20, as well as members of the Bcl-2 family (22, 26). To determine if the targeting motif directed ORFV125 to the mitochondria, we created an N-terminal GFP fusion protein (ORFV125wt) and visualized its subcellular localization by confocal microscopy. HeLa cells transfected with the GFP-only expression vector showed a uniform green fluorescence signal throughout the cytoplasm (Fig. 3C, a), while ORFV125wt-transfected cells revealed a punctate fluorescence around the nucleus (Fig. 3C, d) that coincided with the fluorescent signal of the mitochondrial dye Mitotracker Red (Fig. 3C, e and f). In addition to the full-length construct, we generated a C-terminally truncated protein lacking the putative mitochondrion-targeting sequence (ORFV125Δts) and a construct with only the 30-amino-acid C-terminal targeting motif of ORFV125 fused to GFP (ORFV125ts) (Fig. 3B). The latter construct displayed the same staining pattern as the full-length protein (Fig. 3C, g to i), while the construct lacking the targeting sequence showed a cytoplasmic localization similar to that of GFP only (Fig. 3C, j to l). A further GFP fusion construct (ORFV125AAA) in which the three positively charged amino acids (arginines) downstream of the ORFV125 TM domain were replaced with neutral amino acids (alanines) was observed at mitochondrial and other intracellular membranes (Fig. 3C, m to o).
FIG. 3.
The mitochondrion-targeting motif of ORFV125 is necessary and sufficient to direct the protein to the mitochondria. (A) Comparison of the C-terminal region of ORFV125 with the consensus mitochondrion-targeting motif of tail-anchored proteins (26). The hydrophobic amino acids (aa) constituting the TM domain are boxed. The TM domain is flanked by positively charged amino acids, which are shown as B in the consensus sequence and are in bold in ORFV125. In the consensus sequence, x stands for any amino acid. (B) Schematic representations of GFP fusion constructs. ORFV125wt consists of the full-length 173-amino-acid protein. ORFV125Δts lacks the last 33 amino acids of the protein, which contain the TM domain flanked by the positively charged amino acids. The ORFV125ts construct possesses only the C-terminal 30 amino acids of the viral protein fused to GFP. In ORFV125AAA, the three arginines downstream of the TM domain were changed to alanines. (C) HeLa cells were transiently transfected with the indicated GFP fusion plasmids and visualized by confocal microscopy 24 h later. Mitochondria were labeled with Mitotracker.
We compared the subcellular localization of ORFV125 with that of Bcl-2 and Bcl-w. The green fluorescence of the Bcl-2 fusion protein was visible at the mitochondria, as well as other intracellular membranes (Fig. 3C, p to r), presumably the ER and nuclear membranes, as described by others (18). Bcl-w was localized at the mitochondria but also in the cytoplasm (Fig. 3C, s to u), probably reflecting its overexpression and its loose attachment to the mitochondria (54).
These results show that the mitochondrion-targeting motif of ORFV125 is necessary and sufficient to direct the protein exclusively to the mitochondria and that the basic amino acids downstream of the TM domain contribute to its mitochondrion-specific localization.
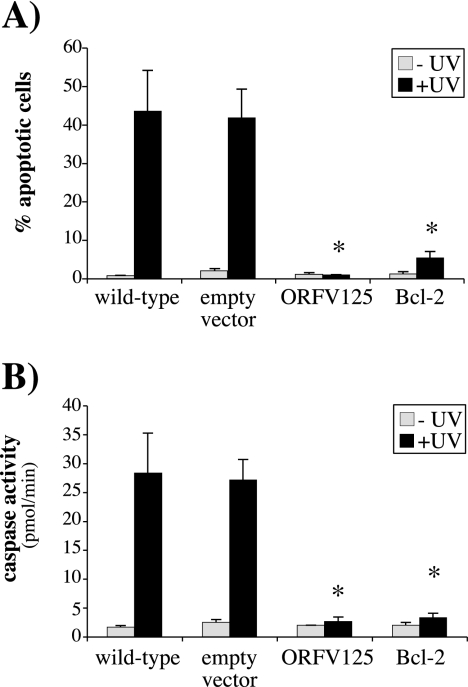
ORFV125 prevents UV-induced apoptosis.
To assess the ability of ORFV125 to inhibit apoptosis, we created 143B cell lines stably expressing ORFV125, Bcl-2, or the empty vector. The DNA content of these cell lines was examined by flow cytometry 17 h after UV irradiation. As shown in Fig. 4A, the wild-type and empty-vector cell lines were susceptible to UV-induced apoptosis but cells expressing ORFV125 or Bcl-2 were fully resistant to UV-induced DNA fragmentation. There was no significant difference in the abilities of ORFV125 and Bcl-2 to inhibit UV-induced apoptosis.
FIG. 4.
ORFV125 inhibits UV-induced apoptosis in the absence of virus infection. (A) Stable 143B cell lines expressing ORFV125, Bcl-2, or the empty vector were UV irradiated. After 17 h, the DNA content of permeabilized, PI-stained cells was analyzed by flow cytometry and the percentage of hypodiploid (apoptotic) cells in each population was calculated. (B) Caspase activity in the indicated cell lines was quantified 7 h after UV treatment. The data shown in panels A and B are averages of three independent experiments with standard deviations indicated. UV-irradiated samples that showed significantly (P < 0.001) less apoptosis or caspase activity than the UV-irradiated wild-type cell line are indicated by asterisks.
As a further test of the antiapoptotic activity of ORFV125, we examined caspase activation in these cell lines. Cells expressing either ORFV125 or Bcl-2 were resistant to UV-induced caspase activation, while the wild-type and empty-vector cell lines showed high levels of caspase activity (Fig. 4B).
Taken together, these results show that ORFV125, without the participation of additional ORFV proteins, can fully inhibit UV-induced DNA fragmentation and caspase activation. Furthermore, its activity is comparable to that of the cellular antiapoptotic protein Bcl-2.
The mitochondrial localization of ORFV125 correlates with apoptosis inhibition.
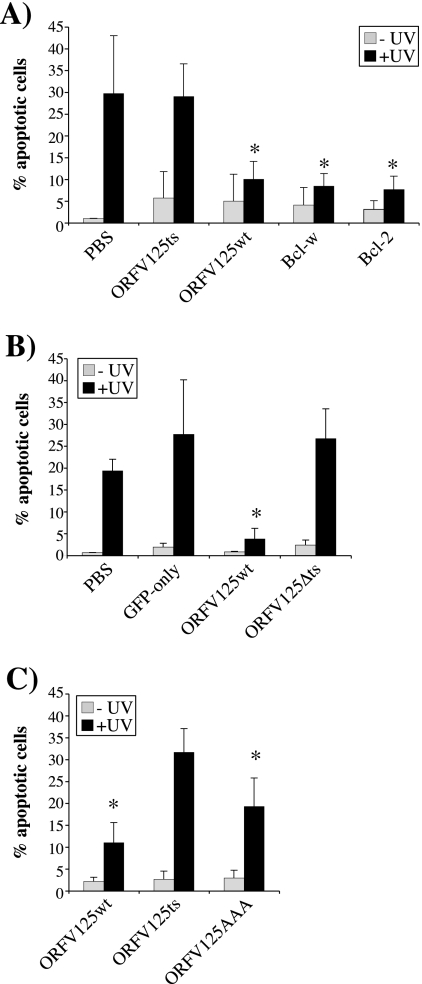
To determine the significance of ORFV125's mitochondrial targeting in its ability to inhibit apoptosis, we examined the abilities of the GFP fusion constructs to prevent UV-induced DNA fragmentation (Fig. 5). Approximately 20 to 30% of the cells either mock transfected (PBS) or transfected with the GFP-only control underwent apoptosis, whereas only 3 to 10% of the cells transfected with the ORFV125wt construct were apoptotic (Fig. 5A and B). Moreover, this level of protection against UV-induced DNA fragmentation was the same as that afforded by the antiapoptotic proteins Bcl-2 and Bcl-w (Fig. 5A). We determined that neither ORFV125 mutant ORFV125ts nor ORFV125Δts was able to prevent apoptosis (Fig. 5A and B), indicating that the mitochondrion-targeting motif is not in itself sufficient to prevent apoptosis and that mitochondrial localization is essential for the protein's function. In support of this last conclusion, we observed that ORFV125AAA, which is only partly localized at the mitochondria, had intermediate antiapoptotic abilities that were less potent than those of ORFV125wt but significantly greater than those of the noninhibitory ORFV125ts mutant (Fig. 5C).
FIG. 5.
The mitochondrial localization of ORFV125 is necessary for its antiapoptotic function. 143B cells were mock transfected (PBS) or transfected with the indicated GFP fusion constructs and UV irradiated either 30 h (A and C) or 12 h (B) later. After a further 17 h (A and C) or 12 h (B), the DNA content of permeabilized, PI-stained cells was examined by flow cytometry. The shortened time between transfection and harvesting in panel B was used to avoid degradation of the ORFV125Δts construct within the cell. The percentage of hypodiploid (apoptotic) cells in each population was calculated. The data presented are averages of three independent experiments with standard deviations indicated. Asterisks indicate UV-irradiated samples in which the apoptotic population is significantly reduced (A, P < 0.05 for ORFV125wt and P < 0.01 for Bcl-w and Bcl-2; B, P < 0.05; C, P < 0.001 for ORFV125wt and P < 0.01 for ORFV125AAA) compared with the corresponding UV-irradiated mock-transfected (A and B) or ORFV125ts-transfected (C) sample.
ORFV125 acts at the mitochondria to inhibit downstream events but has no function upstream of the mitochondria.
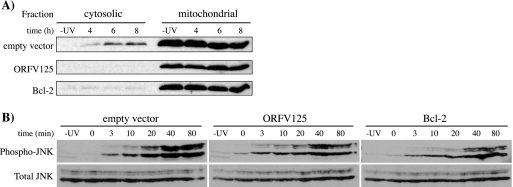
As a direct measurement of the mitochondrial apoptotic pathway, we tested the 143B stable cell lines for UV-induced release of cytochrome c from the mitochondria (Fig. 6A). Four hours after UV treatment, some cytochrome c was detected in the cytoplasm of the empty-vector cell line, with greater amounts detected at 6 and 8 h after UV treatment. Within this time frame, UV irradiation did not cause cytochrome c release in cells expressing ORFV125 or Bcl-2. We also tested the stable cell lines for the ability to prevent phosphorylation of JNK. It has been shown that JNK is activated in response to UV irradiation and that it, in turn, activates Bcl-2 family members (29), which makes it a useful marker of apoptotic responses upstream of mitochondria. Activated JNK was detected in the empty-vector cell line as early as 10 min after UV treatment. Similar results were obtained with the ORFV125 and Bcl-2 cell lines, indicating that neither ORFV125 nor Bcl-2 substantially blocks the phosphorylation of JNK in response to UV treatment (Fig. 6B).
FIG. 6.
ORFV125 inhibits cytochrome c release, but not JNK phosphorylation, following UV treatment. Stable 143B cell lines expressing ORFV125, Bcl-2, or the empty vector were either mock treated (−UV) or UV irradiated. (A) Cells were fractionated into cytosolic and mitochondrial fractions after 4, 6, or 8 h. Release of cytochrome c from the mitochondria into the cytosol was determined by Western blot analysis with an anti-cytochrome c antibody. This blot is representative of three independent experiments. (B) Cell lysates were prepared after the indicated times. Cells harvested at 0 min were collected immediately after UV exposure. Immunoblot analysis was performed to compare the levels of two isozymes of phosphorylated JNK (p46-JNK1 and p54-JNK2) with total JNK. The latter also served as a loading control.
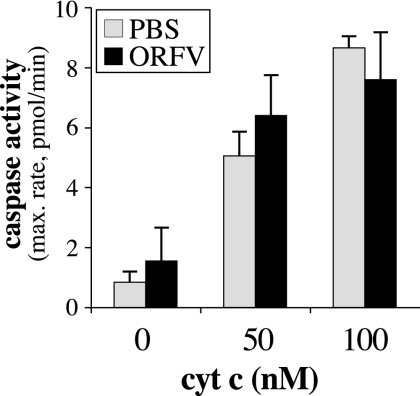
We also examined the possibility that ORFV might express additional factors able to prevent apoptosis downstream of the release of cytochrome c. Cytosolic extracts of ORFV-infected cells were supplemented with different amounts of cytochrome c, and the activation of caspases was then determined (Fig. 7). Mock-infected (PBS-treated) cells showed a dose-dependent increase in caspase activity in response to cytochrome c. The same result was obtained with ORFV-infected cells, suggesting that no ORFV protein within the cytosol can inhibit caspase activation induced by cytosolic cytochrome c.
FIG. 7.
ORFV does not express a cytosolic protein able to inhibit caspase activation induced by cytosolic cytochrome c (cyt c). 143B cells were mock infected (PBS) or infected with ORFV NZ2 (ORFV) at a multiplicity of infection of 5 for 14 h. Cytosolic extracts were prepared and treated with 0, 50, or 100 nM cytochrome c and 1 mM dATP. Activation of caspases was measured in triplicate over 120 min, and the average maximum (max.) rate of AMC production was determined (standard deviations are indicated). The results shown are representative of three independent experiments.
ORFV125 is a Bcl-2-like protein.
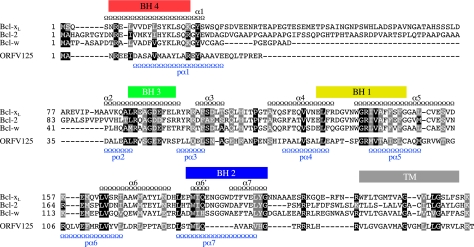
In light of our findings that ORFV125 appears to act in a Bcl-2-like manner, we looked for any evidence of sequence similarities to Bcl-2 family members. Standard BLAST analysis failed to detect homologies between ORFV125 and these proteins (data not shown). However, in addition to its C-terminal, mitochondrion-targeting motif, ORFV125 shares similarities in size and a predicted compact globular structure with Bcl-2 family members. A ClustalW alignment of ORFV125 and three antiapoptotic members of the Bcl-2 family whose solution structures have been resolved, i.e., Bcl-2 (37), Bcl-xL (36), and Bcl-w (21), was conducted to investigate further similarities among these proteins (Fig. 8). This revealed that ORFV125 possesses recognizable BH1 and BH3 sequence motifs and provided some evidence of conserved residues of a BH4 domain and a partial BH2 domain. Furthermore, with secondary-structure prediction programs, we identified seven α-helices in ORFV125 at locations similar to those of the eight α-helices of Bcl-xL, which was used as a representative of the Bcl-2 family (Fig. 8). These observations suggest that ORFV125 might adopt a similar three-dimensional structure. Consistent with that possibility, we found that the HHpred program (45), which compares proteins on the basis of secondary-structure similarities, identified Mcl-1, Bcl-xL, Bcl-2, and Bcl-w as the first four hits.
FIG. 8.
Sequence and predicted secondary-structure similarities between ORFV125 and Bcl-2 family members. Amino acid sequence alignment of ORFV125 with three antiapoptotic Bcl-2 family members, Bcl-xL, Bcl-2, and Bcl-w. BH domains of Bcl-xL are shown as colored boxes, and the TM domain is shown as a gray box. Positions of the Bcl-xL α-helices (α1 to -6, -6′, and -7) (Jpred) are represented as black chains above the alignment, while the predicted α-helices for ORFV125 (pα1 to -7) are plotted below the alignment in blue. Amino acids conserved in all four sequences are marked with black boxes showing amino acids identical to ORFV125 and gray boxes indicating conserved substitutions (according to ClustalW).
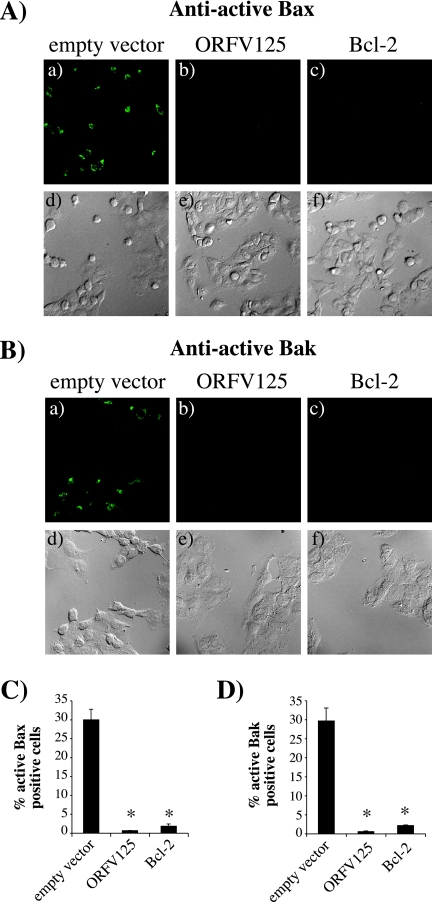
One function of antiapoptotic Bcl-2 family members is to prevent the activation of the proapoptotic mediators Bax and Bak upon an apoptotic stimulus. Their activation is initiated by a conformational change which is characterized by exposure of the N terminus and can be detected with antibodies specifically recognizing this N-terminal epitope (8). To determine whether ORFV125 inhibits apoptosis in a Bcl-2-like manner, we examined the activation of Bax and Bak in UV-treated 143B stable cell lines (Fig. 9). While 30% of the cells in the empty-vector control contained active Bax or Bak, cells expressing ORFV125 and Bcl-2 did no show any active protein. Thus, like Bcl-2, ORFV125 inhibited the UV-induced activation of Bax and Bak.
FIG. 9.
ORFV125 inhibits the activation of Bax and Bak. 143B stable cell lines were incubated with 50 μM Z-VAD-FMK (Calbiochem), added 1 h before UV treatment. Eight hours after treatment, cells were stained with Bax (A) or Bak (B) conformation-specific antibodies and visualized by fluorescence (a to c) or phase-contrast (d to f) microscopy. The percentage of cells with active Bax (C) or Bak (D) in four different microscope fields was calculated and averaged. The results are presented as an average of three independent experiments (standard deviations are indicated). Samples that showed significantly (P < 0.001) less active Bax or Bak than the empty-vector control cell line are indicated by asterisks.
DISCUSSION
Among the array of host modulating strategies deployed by poxviruses, a common theme is the inhibition of apoptosis of infected cells. A substantial number of different poxviral antiapoptotic proteins have been identified, and it was therefore somewhat surprising that inspection of the genome of ORFV revealed little evidence of such inhibitors of apoptosis. In this study, we set out to test the abilities of ORFV to inhibit apoptosis and to identify the ORFV gene(s) responsible for any such activity. Our results show that ORFV expresses potent apoptotic inhibitory activity, and we were able to identify a single gene that is at least in part, or perhaps fully, responsible for that activity. The gene (ORFV125) encodes a protein that is directed to the mitochondria by a C-terminal domain which has a strong match with the consensus sequence of the C-terminal signal motif of mitochondrion-targeted proteins (26). Consistent with the mitochondrial location of ORFV125, we were able to show that in cells subjected to an apoptotic stimulus, the viral protein inhibits the release of cytochrome c, caspase activation, and DNA fragmentation. However, it did not inhibit JNK activation, which occurs at a step in the apoptotic response cascade upstream of the mitochondria. Nor could we detect evidence of any cytosol-localized ORFV factors able to inhibit caspase activation downstream of the release of cytochrome c from the mitochondria.
ORFV125's specific localization to the mitochondria seems to be necessary for its antiapoptotic function, as a deletion of the C-terminal targeting domain led to the distribution of the truncated protein throughout the cytoplasm and a loss of inhibitory activity. In addition, a mutant form of ORFV125 (ORFV125AAA), in which the positively charged amino acids downstream of the TM domain were converted to neutral amino acids, showed only partial localization to the mitochondria and a similar partial loss of inhibitory activity. The partial retention of this mutant protein within the mitochondria was unexpected as a similar mutant of F1L, a mitochondrial apoptosis inhibitor of VACV, was redirected entirely to the ER (46). The behavior of ORFV125AAA may reflect the unusually high basicity of the 11-amino-acid region immediately upstream of the TM domain. This region contains five arginine and two histidine residues, whereas the corresponding region of VACV F1L contains only two basic residues. This partial retention within the mitochondria can also be seen with a Bcl-xL mutant similar to the ORF125AAA construct, which is localized to all intracellular membranes, whereas wild-type Bcl-xL is directed only to the mitochondria (26).
We showed that the ability of ORFV125 to inhibit UV-induced apoptosis was comparable to that of Bcl-2. Although the overall amino acid sequence identity of these two proteins is only 10%, the alignment of ORFV125 and three antiapoptotic members of the Bcl-2 family revealed that ORFV125 shares predicted structural features and key functional residues with Bcl-2 proteins, including BH domains 1 and 3, as well as partial evidence of BH2 and BH4 domains. Crystallization studies of Bcl-xL have revealed residues within the hydrophobic groove formed by the BH1, BH2, and BH3 domains that interact with BH3 peptides of proapoptotic proteins such as Bak, Bad, and Bim (30, 38, 43). Several of these residues are conserved in ORFV125, suggesting that the viral protein might interact with proapoptotic family members and inhibit apoptosis in a manner similar to that used by antiapoptotic Bcl-2 proteins. These observations indicate that ORFV125 can be classified as a viral Bcl-2-like protein.
This is the first analysis of a parapoxvirus inhibitor of apoptosis. Orthologs of ORFV125 are encoded by each of the recognized species of parapoxviruses, with interspecies amino acid sequence identities ranging from 60 to 80% (7; unpublished observations). Each ortholog displays the Bcl-2-like features described for ORFV125.
We also compared ORFV125 with other mitochondrion-localized antiapoptotic proteins of the poxvirus family, i.e., FWPV039, which is a recognized Bcl-2 homolog, and M11L and F1L, which have not been regarded as Bcl-2 family members although BH3-like domains have been detected in both proteins (40, 50). The amino acid sequence identity of these four proteins is only approximately 10%. Nevertheless, they are similar in size, apart from an N-terminal extension in F1L, and each protein possesses a C-terminal mitochondrion-targeting motif (Fig. 10). Beyond that, ORFV125 and FWPV039 exhibit at least two of four BH domains (1), and within the same regions of M11L and F1L one can find residues consistent with the consensus sequences of BH1 and BH3 domains (Fig. 10). The region of F1L aligned with the BH3 consensus motif differs from the potential BH3-like domain identified by Postigo et al. (40). Figure 10 also shows that within each genus of the vertebrate poxvirus subfamily, apart from the Molluscipoxvirus genus, one can identify a corresponding protein that is of a similar size, exhibits a potential mitochondrion-targeting motif, and displays evidence of shared sequence within the potential BH1- and BH3-like domains. We excluded the protein MC008L of molluscum contagiosum virus from our alignment, as it does not exhibit discernible BH-like domains although it shares the other Bcl-2-like features. In general, this group of proteins displays only limited overall sequence similarities. However, substantial similarities are apparent within the proposed BH-like domains, suggesting that these regions are functionally significant. We therefore suggest that the majority of the vertebrate poxvirus subfamily members encode proteins that display a spectrum of Bcl-2-like properties and are likely to be functional homologs of the Bcl-2 family.
FIG. 10.
Representatives from seven of the eight vertebrate poxvirus genera encode proteins with Bcl-2-like features. Alignment of BH3-like and BH1-like domains of proteins from fowlpox virus (FWPV, FWPV039), ORFV (ORFV125), myxoma virus (MYXV, M11L), sheeppox virus (SPPV, SPPV014), swinepox virus (SWPV, SWPV012), Yaba monkey tumor virus (YMTV, 16L), deerpox virus (DPV, DPV022), and VACV (F1L). Deerpox virus is an unassigned member of the subfamily. Amino acids highlighted in yellow match the consensus sequence of the BH1 domain (Prosite accession no. PSO1080) or the BH3 domain (Prosite accession no. PSO1259). The Prosite consensus sequences are shown at the top, with X representing any amino acid. Amino acids boxed in blue are conserved in at least five of eight poxviral proteins. Predicted mitochondrion-targeting sequences (mTS) are marked as red boxes. xn indicates the numbers of intervening amino acids. The total number of amino acids in each protein is shown at the end of each sequence.
Functional analysis has only been reported for M11L and F1L, which have been shown to act similarly to antiapoptotic Bcl-2 proteins. M11L constitutively interacts with Bak (50) but only binds to Bax following an apoptotic stimulus (47). F1L also binds to Bak constitutively (40, 51), but instead of interacting with Bax, it sequesters the BH3-only protein BimL (48). In addition, the crystallographic structure of M11L has recently been reported and it confirms the Bcl-2-like nature of the viral protein (27).
The precise mechanism of action of ORFV125 still needs to be clarified. Its ability to inhibit the activation of Bax and Bak following UV treatment indicates that ORFV125 shares functional features with M11L and F1L. However, the low sequence similarity between the mitochondrion-localized poxviral proteins suggests that ORFV125 may well have functional characteristics that differ from those of M11L and F1L. Further experiments are under way to determine if ORFV125's ability to block the activation of Bax and Bak is due to direct interactions with these proteins or the sequestering of other proteins, such as BH3-only family members, away from Bax and Bak.
In summary, our results show that ORFV125 is a potent Bcl-2-like protein that inhibits UV-induced apoptosis without the participation of other ORFV proteins. Poxviruses commonly encode multiple inhibitors of apoptosis, but we have not been able to identify any additional antiapoptotic proteins encoded by ORFV. In addition, we have not been able to create a stable ORFV125 deletion virus, suggesting that this protein is essential for infection, as might be the case if it were the only ORFV apoptotic inhibitor. This possibility and the proposal that almost all vertebrate poxviruses possess similar mitochondrial inhibitors of apoptosis illustrate the importance of mitochondria as a control point in the apoptotic pathways.
Acknowledgments
This work was supported by the Health Research Council of New Zealand, the New Economy Research Fund (NERF), and the University of Otago.
We thank Catherine Day (University of Otago) for helpful discussions, reading the manuscript, and providing reagents. We are also grateful to David Huang (The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia) for providing a range of plasmids, Andrew McNaughton for assistance with confocal microscopy, Gill Hughes for technical support, and Trudie Bateman and Alison Mercer for assistance with the results shown in Fig. 1A.
Footnotes
Published ahead of print on 2 May 2007.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 3.Breckenridge, D. G., and D. Xue. 2004. Regulation of mitochondrial membrane permeabilization by BCL-2 family proteins and caspases. Curr. Opin. Cell Biol. 16:647-652. [DOI] [PubMed] [Google Scholar]
- 4.Brick, D. J., R. D. Burke, A. A. Minkley, and C. Upton. 2000. Ectromelia virus virulence factor p28 acts upstream of caspase-3 in response to UV light-induced apoptosis. J. Gen. Virol. 81:1087-1097. [DOI] [PubMed] [Google Scholar]
- 5.Cartier, J. L., P. A. Hershberger, and P. D. Friesen. 1994. Suppression of apoptosis in insect cells stably transfected with baculovirus p35: dominant interference by N-terminal sequences p351-76. J. Virol. 68:7728-7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuconati, A., and E. White. 2002. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 16:2465-2478. [DOI] [PubMed] [Google Scholar]
- 7.Delhon, G., E. R. Tulman, C. L. Afonso, Z. Lu, A. de la Concha-Bermejillo, H. D. Lehmkuhl, M. E. Piccone, G. F. Kutish, and D. L. Rock. 2004. Genomes of the parapoxviruses Orf virus and bovine papular stomatitis virus. J. Virol. 78:168-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desagher, S., and J. C. Martinou. 2000. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 10:369-377. [DOI] [PubMed] [Google Scholar]
- 9.Dix, B. R., S. J. O'Carroll, C. J. Myers, S. J. Edwards, and A. W. Braithwaite. 2000. Efficient induction of cell death by adenoviruses requires binding of E1B55k and p53. Cancer Res. 60:2666-2672. [PubMed] [Google Scholar]
- 10.Everett, H., M. Barry, S. F. Lee, X. Sun, K. Graham, J. Stone, R. C. Bleackley, and G. McFadden. 2000. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J. Exp. Med. 191:1487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, H., and G. McFadden. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160-165. [DOI] [PubMed] [Google Scholar]
- 12.Everett, H., and G. McFadden. 2002. Poxviruses and apoptosis: a time to die. Curr. Opin. Microbiol. 5:395-402. [DOI] [PubMed] [Google Scholar]
- 13.Falkner, F. G., S. Chakrabarti, and B. Moss. 1987. pUV I: a new vaccinia virus insertion and expression vector. Nucleic Acids Res. 15:7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming, S. B., and A. A. Mercer. 2006. Genus Parapoxvirus, p. 127-165. In A. A. Mercer, A. Schmidt, and O. Weber (ed.), Poxviruses, 1st ed. Birkhäuser Verlag, Basel, Switzerland.
- 15.Garrido, C., L. Galluzzi, M. Brunet, P. E. Puig, C. Didelot, and G. Kroemer. 2006. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 13:1423-1433. [DOI] [PubMed] [Google Scholar]
- 16.Green, D. R., and G. Kroemer. 2004. The pathophysiology of mitochondrial cell death. Science 305:626-629. [DOI] [PubMed] [Google Scholar]
- 17.Gurudutta, G. U., Y. K. Verma, V. K. Singh, P. Gupta, H. G. Raj, R. K. Sharma, and R. Chandra. 2005. Structural conservation of residues in BH1 and BH2 domains of Bcl-2 family proteins. FEBS Lett. 579:3503-3507. [DOI] [PubMed] [Google Scholar]
- 18.Häcki, J., L. Egger, L. Monney, S. Conus, T. Rosse, I. Fellay, and C. Borner. 2000. Apoptotic crosstalk between the endoplasmic reticulum and mitochondria controlled by Bcl-2. Oncogene 19:2286-2295. [DOI] [PubMed] [Google Scholar]
- 19.Hampton, M. B., B. Zhivotovsky, A. F. Slater, D. H. Burgess, and S. Orrenius. 1998. Importance of the redox state of cytochrome c during caspase activation in cytosolic extracts. Biochem. J. 329(Pt. 1):95-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardwick, J. M., and D. S. Bellows. 2003. Viral versus cellular BCL-2 proteins. Cell Death Differ 10(Suppl. 1):S68-S76. [DOI] [PubMed] [Google Scholar]
- 21.Hinds, M. G., M. Lackmann, G. L. Skea, P. J. Harrison, D. C. Huang, and C. L. Day. 2003. The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity. EMBO J. 22:1497-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie, C., H. Suzuki, M. Sakaguchi, and K. Mihara. 2002. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell 13:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, D. C., S. Cory, and A. Strasser. 1997. Bcl-2, Bcl-XL and adenovirus protein E1B19kD are functionally equivalent in their ability to inhibit cell death. Oncogene 14:405-414. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, G., M. P. Murphy, and E. C. Ledgerwood. 2005. Mitochondrial reactive oxygen species regulate the temporal activation of nuclear factor κB to modulate tumour necrosis factor-induced apoptosis: evidence from mitochondria-targeted antioxidants. Biochem. J. 389:83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, Z., and W. S. El-Deiry. 2005. Overview of cell death signaling pathways. Cancer Biol. Ther. 4:139-163. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann, T., S. Schlipf, J. Sanz, K. Neubert, R. Stein, and C. Borner. 2003. Characterization of the signal that directs Bcl-xL, but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kvansakul, M., M. F. van Delft, E. F. Lee, J. M. Gulbis, W. D. Fairlie, D. C. Huang, and P. M. Colman. 2007. A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak. Mol. Cell 25:933-942. [DOI] [PubMed] [Google Scholar]
- 28.Lawen, A. 2003. Apoptosis—an introduction. Bioessays 25:888-896. [DOI] [PubMed] [Google Scholar]
- 29.Lei, K., and R. J. Davis. 2003. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA 100:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, X., S. Dai, Y. Zhu, P. Marrack, and J. W. Kappler. 2003. The structure of a Bcl-xL/Bim fragment complex: implications for Bim function. Immunity 19:341-352. [DOI] [PubMed] [Google Scholar]
- 31.Liu, X., C. N. Kim, J. Yang, R. Jemmerson, and X. Wang. 1996. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell 86:147-157. [DOI] [PubMed] [Google Scholar]
- 32.Maguire, T., P. Harrison, O. Hyink, J. Kalmakoff, and V. K. Ward. 2000. The inhibitors of apoptosis of Epiphyas postvittana nucleopolyhedrovirus. J. Gen. Virol. 81:2803-2811. [DOI] [PubMed] [Google Scholar]
- 33.McInnes, C. J., A. R. Wood, and A. A. Mercer. 1998. Orf virus encodes a homolog of the vaccinia virus interferon-resistance gene E3L. Virus Genes 17:107-115. [DOI] [PubMed] [Google Scholar]
- 34.Mercer, A. A., N. Ueda, S. M. Friederichs, K. Hofmann, K. M. Fraser, T. Bateman, and S. B. Fleming. 2006. Comparative analysis of genome sequences of three isolates of Orf virus reveals unexpected sequence variation. Virus Res. 116:146-158. [DOI] [PubMed] [Google Scholar]
- 35.Mercer, A. A., D. L. Yirrell, E. M. Whelan, P. F. Nettleton, I. Pow, J. A. Gilray, H. W. Reid, and A. J. Robinson. 1997. A novel strategy for determining protective antigens of the parapoxvirus, Orf virus. Virology 229:193-200. [DOI] [PubMed] [Google Scholar]
- 36.Muchmore, S. W., M. Sattler, H. Liang, R. P. Meadows, J. E. Harlan, H. S. Yoon, D. Nettesheim, B. S. Chang, C. B. Thompson, S. L. Wong, S. L. Ng, and S. W. Fesik. 1996. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 381:335-341. [DOI] [PubMed] [Google Scholar]
- 37.Petros, A. M., A. Medek, D. G. Nettesheim, D. H. Kim, H. S. Yoon, K. Swift, E. D. Matayoshi, T. Oltersdorf, and S. W. Fesik. 2001. Solution structure of the antiapoptotic protein bcl-2. Proc. Natl. Acad. Sci. USA 98:3012-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petros, A. M., D. G. Nettesheim, Y. Wang, E. T. Olejniczak, R. P. Meadows, J. Mack, K. Swift, E. D. Matayoshi, H. Zhang, C. B. Thompson, and S. W. Fesik. 2000. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 9:2528-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petros, A. M., E. T. Olejniczak, and S. W. Fesik. 2004. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta 1644:83-94. [DOI] [PubMed] [Google Scholar]
- 40.Postigo, A., J. R. Cross, J. Downward, and M. Way. 2006. Interaction of F1L with the BH3 domain of Bak is responsible for inhibiting vaccinia-induced apoptosis. Cell Death Differ. 13:1651-1662. [DOI] [PubMed] [Google Scholar]
- 41.Robinson, A. J., G. Ellis, and T. Balassu. 1982. The genome of Orf virus: restriction endonuclease analysis of viral DNA isolated from lesions of Orf in sheep. Arch. Virol. 71:43-55. [DOI] [PubMed] [Google Scholar]
- 42.Robinson, A. J., and A. A. Mercer. 1988. Orf virus and vaccinia virus do not cross-protect sheep. Arch. Virol. 101:255-259. [DOI] [PubMed] [Google Scholar]
- 43.Sattler, M., H. Liang, D. Nettesheim, R. P. Meadows, J. E. Harlan, M. Eberstadt, H. S. Yoon, S. B. Shuker, B. S. Chang, A. J. Minn, C. B. Thompson, and S. W. Fesik. 1997. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275:983-986. [DOI] [PubMed] [Google Scholar]
- 44.Schinzel, A., T. Kaufmann, and C. Borner. 2004. Bcl-2 family members: intracellular targeting, membrane-insertion, and changes in subcellular localization. Biochim. Biophys. Acta 1644:95-105. [DOI] [PubMed] [Google Scholar]
- 45.Söding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 46.Stewart, T. L., S. T. Wasilenko, and M. Barry. 2005. Vaccinia virus F1L protein is a tail-anchored protein that functions at the mitochondria to inhibit apoptosis. J. Virol. 79:1084-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su, J., G. Wang, J. W. Barrett, T. S. Irvine, X. Gao, and G. McFadden. 2006. Myxoma virus M11L blocks apoptosis through inhibition of conformational activation of Bax at the mitochondria. J. Virol. 80:1140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, J. M., D. Quilty, L. Banadyga, and M. Barry. 2006. The vaccinia virus protein F1L interacts with Bim and inhibits activation of the pro-apoptotic protein Bax. J. Biol. Chem. 281:39728-39739. [DOI] [PubMed] [Google Scholar]
- 49.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2004. The genome of canarypox virus. J. Virol. 78:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, G., J. W. Barrett, S. H. Nazarian, H. Everett, X. Gao, C. Bleackley, K. Colwill, M. F. Moran, and G. McFadden. 2004. Myxoma virus M11L prevents apoptosis through constitutive interaction with Bak. J. Virol. 78:7097-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wasilenko, S. T., L. Banadyga, D. Bond, and M. Barry. 2005. The vaccinia virus F1L protein interacts with the proapoptotic protein Bak and inhibits Bak activation. J. Virol. 79:14031-14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wasilenko, S. T., T. L. Stewart, A. F. Meyers, and M. Barry. 2003. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc. Natl. Acad. Sci. USA 100:14345-14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willis, S. N., and J. M. Adams. 2005. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17:617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson-Annan, J., L. A. O'Reilly, S. A. Crawford, G. Hausmann, J. G. Beaumont, L. P. Parma, L. Chen, M. Lackmann, T. Lithgow, M. G. Hinds, C. L. Day, J. M. Adams, and D. C. Huang. 2003. Proapoptotic BH3-only proteins trigger membrane integration of prosurvival Bcl-w and neutralize its activity. J. Cell Biol. 162:877-887. [DOI] [PMC free article] [PubMed] [Google Scholar]