Abstract
Three transgenic rabbit lines that express a well-characterized human major histocompatibility complex class I (MHC-I) gene (HLA-A2.1) have been established. All three lines carry the HLA-A2.1 heavy chain and are able to pass the transgene to their offspring with both the outbred and the inbred EIII/JC genetic background. HLA-A2.1 colocalizes exclusively with rabbit MHC-I on the cell surfaces. These HLA-A2.1 transgenic rabbits demonstrated infection patterns similar to those found after cottontail rabbit papillomavirus (CRPV) challenge when compared with results in normal rabbits, although higher regression rates were found in HLA-A2.1 transgenic rabbits. Because the CRPV genome can accommodate significant modifications, the CRPV/HLA-A2.1 rabbit model has the potential to be used to screen HLA-A2.1-restricted immunogenic epitopes from human papillomaviruses in the context of in vivo papillomavirus infection.
Human papillomaviruses (HPVs) are strongly correlated with various human cancers, in particular, cervical cancer and head and neck cancer. Recent advances in HPV vaccine development have led to a protective viruslike particle (VLP) vaccine for clinical use (7, 11, 29, 32). However, because of the type-specific nature of the protective antibody responses to these viruses, a vaccine that can elicit more broadly protective neutralizing antibodies will be required for more complete protection against HPV-associated cancer (31). In addition, the VLP vaccine only provides protection against new infections and is expected to have little or no impact on existing HPV disease (25). To eliminate existing HPV infections, a second form of immunity, namely cell-mediated immunity, needs to be activated (27).
The extreme species specificity of papillomaviruses prevents the use of immunocompetent laboratory animals to study HPV infections (8). Current preclinical models of natural papillomavirus infections include rabbit, dog, and bovine models (3). The cottontail rabbit papillomavirus (CRPV) rabbit model offers several advantages as a preclinical model for studying host immunity to papillomavirus infection (1, 4). The model has been used extensively to study protective immunity to VLP-based vaccines, as well as cell-mediated immunity to various viral early proteins, including E1, E2, E6, E7, E8, and L1 (13, 17, 24). A powerful advantage of the CRPV model is that papillomas can be generated by direct infection of the skin with viral DNA in the absence of encapsidation by the viral coat proteins (2, 16, 22). This observation provides opportunities to genetically alter the virus by site-directed mutagenesis and to engineer epitopes into the various viral genes for testing specific immunities. As an example of this technology, we have shown that an HPV type 16 (HPV-16) E7 epitope can be engineered into the CRPV E7 gene within the CRPV genome. This hybrid genome retains the ability to initiate papillomas (15). Despite these advantages, studies on viral immunity to CRPV in the context of rabbit major human histocompatibility complex (MHC) molecules provide little useful information for the design, induction, and testing of antigen-specific T-cell responses to HPV epitopes in the context of MHC molecules.
Here we report the initial characterization of and studies on our recently established HLA-A2.1 transgenic rabbit model (19). We have demonstrated by Southern blot analysis that the HLA-A2.1 gene is integrated into the rabbit genome; we have also demonstrated similar expression patterns of HLA-A2.1 and rabbit MHC class I (MHC-I) in three rabbit founder lines and their offspring. Furthermore, we have tested CRPV infection in these HLA-A2.1 transgenic rabbits with the use of different CRPV strains. Our data clearly show that HLA-A2.1 transgenic rabbits show a susceptibility to CRPV infection akin to that of normal domestic rabbits.
MATERIALS AND METHODS
Generation of transgenic rabbits.
The procedures for producing transgenic rabbits have been previously described (28). New Zealand White (NZW) rabbits were purchased from Covance Research Products, Inc. (Denver, PA). Female donor rabbits were injected with 120 U of pregnant mare's serum gonadotropin and 150 U of human chorionic gonadotropin to induce superovulation. The rabbits were mated with a fertile male on day 4 immediately after the administration of pregnant mare's serum gonadotropin. Fertilized eggs were collected for microinjection with an HLA-A2.1 DNA construct (3 μg/ml) immediately following the collection. The injected eggs were incubated for 1 h following microinjection and then transplanted into the oviducts of pseudopregnant recipient rabbits that had been mated with a sterile (vasectomized) male at the same time that the donors were mated. The F1 transgenic rabbits used in the experiments were produced by breeding the founder rabbits to NZW rabbits as well as an inbred rabbit line (EIII/JC). NZW rabbits and inbred EIII/JC rabbits were maintained in the animal facility of the Pennsylvania State University College of Medicine. The studies were approved by the Institutional Animal Care and Use Committee of the Pennsylvania State University College of Medicine.
Southern blot analysis.
Transgenic rabbits were identified by Southern blot analysis of DNA isolated from skin biopsy samples. In brief, 10-μg samples of total rabbit genomic DNA were digested with EcoRI in an overnight digestion. The digestions were concentrated in a Microcon 30 (Millipore Corporation) and loaded into an 0.8% agarose gel. The gel was run at 100 V for 2 h, depurinated in 0.1 N HCl for 10 min, denatured two times for 15 min in 1.5 M NaCl, 0.5 M NaOH, and neutralized for 1 h in 1 M NaCl, 0.5 M Tris buffer (pH 7.0). The gel was blotted to Hybond N+ membrane (Amersham) in an overnight transfer. DNA was fixed to the membrane by using a UV Stratalinker 1800 (Stratagene). The HLA-A2.1 heavy chain gene (7.9 kb) was isolated from its parent plasmid (a kind gift from Victor Engelhard) by EcoRI digestion and was labeled with [32P]ATP. Serial dilutions of the unlabeled HLA-A2.1 heavy chain gene were used on the Southern blots as positive size controls. The membranes were prehybridized in Amersham Rapid-Hyb buffer for 5 h at 65°C, and the denatured probe was then added. The membranes were hybridized overnight, and then washed twice for 15 min each with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate at room temperature and twice for 30 min each with 0.1× SSC, 0.1% sodium dodecyl sulfate at 65°C. The membranes were wrapped and exposed to Kodak XOMAT film overnight at −70°C using two intensifying screens.
Immunohistochemistry.
Rabbit ear punch biopsy samples or tissues were collected and frozen in liquid nitrogen and subsequently kept at −70°C. Eight-micrometer cryosections were captured onto glass slides and fixed in cold acetone for 15 min and then blocked by immersing in 0.3% aqueous hydrogen peroxide for 5 min. After being thoroughly rinsed in phosphate-buffered saline (PBS), the slides were incubated in normal horse serum for 30 min (diluted in 3% bovine serum albumin-PBS). The slides were then incubated with the HLA-A2.1-specific monoclonal antibody BB7.2 (ATCC) or mouse anti-rabbit MHC-I (monoclonal antibody 73.2; Spring Valley, Baltimore, MD) for 1 h and with biotinylated secondary antibody for 30 min. After being thoroughly washed, the slides were incubated in an avidin-biotin complex for 30 min as directed in the instructions for the Vector Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, CA). After being washed in PBS three times, the tissues were incubated with aminoethylcarbazole chromogen solution (Zymed AEC substrate chromogen kit; South San Francisco, CA) for 15 to 20 min. Mayer's hematoxylin was used to counterstain the tissues for 5 min. The slides were mounted and covered with coverslips and then checked by bright-field microscopy. The images were taken and recorded digitally. The expression levels of HLA-A2.1, rabbit MHC-I, and CRPV L1 were examined and determined subjectively. The highest levels observed were scaled as “+++” and the lowest levels as “+.” A lack of signal was designated as “−” or negative.
ELISA.
CRPV VLPs generated in our laboratory (1 μg/well in 1× PBS buffer) were bound to 96-well enzyme-linked immunosorbent assay (ELISA) plates (Evergreen Scientific) at room temperature for 30 min, washed, and blocked for 1 h with 1× PBS buffer containing 5% nonfat milk protein as a blocking buffer. Rabbit serum was diluted in 1× PBS blocking buffer and incubated for 1 h, followed by incubation with swine anti-rabbit alkaline phosphatase-conjugated antibody (1:1,000; DAKO) and detection with 1 mg/ml of ρ-nitrophenyl phosphate substrate. Wells probed with normal rabbit sera were used as a negative control. The absorbance at 405 nm (reference filter, 450 nm) was measured with an OPSYS MR microplate reader (Thermo Labsystems).
Flow cytometry.
Aliquots of 106 peripheral blood lymphocytes (PBLs) were harvested from blood as described previously (18) and incubated with 100 μl BB7.2 or a 1:50 dilution of mouse anti-rabbit MHC-I (monoclonal antibody 73.2) supernatant on ice for 45 min. The cells were washed three times with PBS containing 2% fetal bovine serum and then incubated with mouse immunoglobulin G-phycoerythrin-conjugated antibody (1:50) for 45 min. The cells were then washed three times and resuspended in freshly prepared 2% paraformaldehyde, PBS buffer for sorting in a fluorescence-activated cell sorter (FACScan; Becton Dickinson). The control cells were treated using the same protocol, except that the primary antibody incubation step was eliminated. The expression levels were determined by using Cellquest analysis software. The mean fluorescence intensity was recorded for each rabbit and used for statistical analyses. The statistical significance was determined by unpaired t test comparison (P < 0.05 was considered significant).
Virus and viral DNA infection of rabbit skin.
Rabbits were sedated with ketamine at 40 mg/kg of body weight and 5 mg/kg xylazine prior to infection. After shaving and scarification, rabbit back skin was challenged with 50 μl infectious virus (Hershey CRPVp stock; 1:100). For viral DNA infection, the back skin was first wounded with a scalpel blade. Three days later, the rabbits were again sedated, and the scarified sites were scratched with a 21-gauge needle and 10 μg viral DNA (H.CRPVp or H.CRPVr) was applied to the sites (15). Beginning at 3 weeks after virus or viral DNA challenge, the rabbits were monitored for papilloma development. The papilloma size was determined by calculating the cubic root of the products of length × width × height of individual papillomas in millimeters to obtain a geometric mean diameter. The data were represented as the means (± standard errors of the means) of the geometric mean diameters for each test group. The statistical significance was determined by the unpaired t test comparison (P < 0.05 was considered significant).
RESULTS
Presence of HLA-A2.1 gene in rabbit genome shown by Southern blotting.
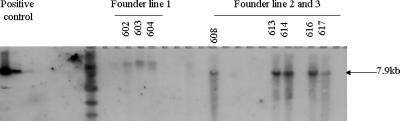
The Southern blot results identified three founder rabbits. Founder line 1 showed a higher-molecular-weight band (about 16 kb), while the other two founder lines showed products of the expected size (7.9 kb; Fig. 1), similar to those in the control lanes on the left of Fig. 1.
FIG. 1.
Integration of HLA-A2.1 in offspring from three rabbit founder lines, as shown by Southern blotting. The positive control lanes (on the left) with different amounts of the HLA-A2.1 heavy chain gene show a 7.9-kb band. Founder line 1 shows a molecular-weight band higher than the predicted size (7.9 kb), while founder lines 2 and 3 show bands of the size anticipated for the integrated transgene.
HLA-A2.1 and MHC-I expression in transgenic rabbits.
As noted above, one founder rabbit contained an HLA-A2.1 gene with a higher-molecular-weight band than those in the other two founders. We have previously demonstrated colocalization of HLA-A2.1 and rabbit MHC-I on different cell surfaces (19). Here, we tested whether HLA-A2.1 expression levels differed on the cell surfaces of these different founder rabbits and also if HLA-A2.1 expression interfered with rabbit MHC-I expression on the cell surfaces. We established one HLA-A2.1 transgenic line (up to F7) from founder line 1 and another HLA-A2.1 transgenic line (up to F6) from cross breeding with founder lines 2 and 3 based on an inbred EIII/JC genetic background. The data presented below are derived from rabbits of these two established transgenic rabbit lines.
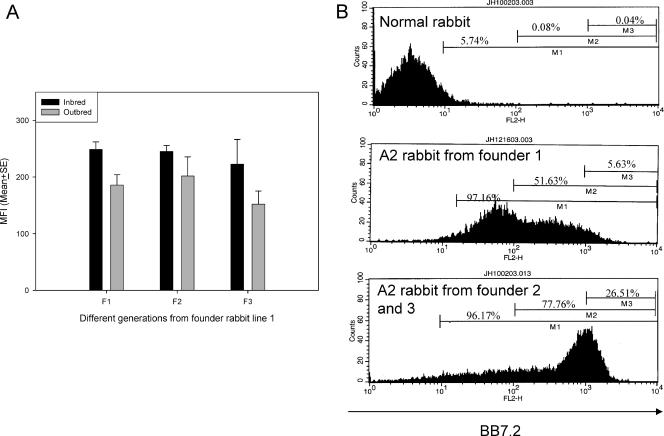
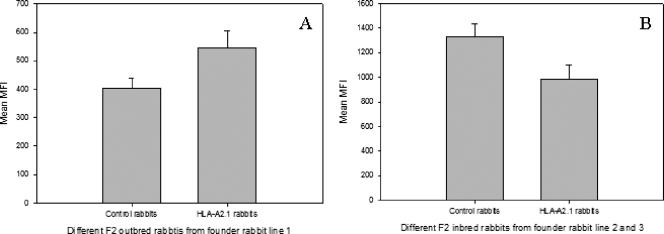
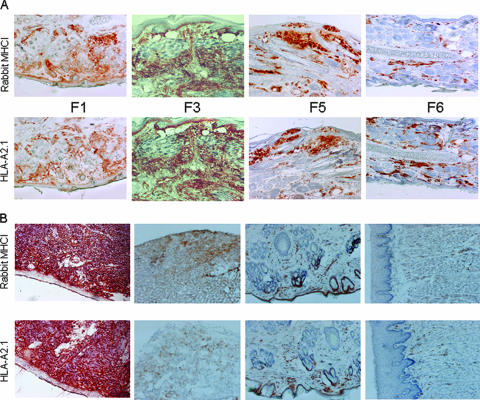
First, we tested the expression levels of HLA-A2.1 and rabbit MHC-I on PBL surfaces by one-color flow cytometry. PBLs separated from normal and HLA-A2.1 transgenic rabbits' blood were labeled with specific anti-rabbit MHC-I or anti-HLA-A2.1 monoclonal antibodies, respectively. The mean fluorescence intensities were recorded. All founder rabbits showed strong HLA-A2.1 expression, despite the differences in DNA sizes in the Southern blot results. To passage the transgene, we bred the founder rabbits with outbred NZW rabbits and inbred EIII/JC rabbits. The HLA-A2.1 expression levels did not change in the offspring from all of these founder lines (Fig. 2A). The offspring from founder lines 2 and 3 showed higher expression levels of HLA-A2.1 than those of founder line 1 (Fig. 2B). The expression levels of rabbit MHC-I, however, were similar in the normal and all transgenic rabbits, even though different expression levels were observed from different labeling experiments (unpaired Student's t test; P > 0.05) (Fig. 3A and B). Second, we tested the in situ expressionlevels of HLA-A2.1 and rabbit MHC-I in rabbit skin tissues. Ear biopsy samples collected from different generations of offspring from these three founder rabbits were examined via immunohistochemistry. Different HLA-A2.1 expression levels were found from rabbit to rabbit, but no significant difference was found among the offspring of these three founder lines. In addition, similar patterns of HLA-A2.1 and MHC-I expression were observed in ear tissues, from the founder rabbits to the descendants (up to F7) (Fig. 4A).
FIG. 2.
(A) Expression levels of HLA-A2.1 on the cell surfaces of PBLs from different generations of rabbit founder line 1. F1 to F3 offspring from founder rabbit line 1 were tested for HLA-A2.1 expression on PBL surfaces. Rabbits from both outbred and inbred genetic backgrounds showed stable levels of HLA-A2.1 from generation to generation. MFI, mean fluorescence intensity; SE, standard error. (B) Representative flow cytometry data from PBLs from a normal rabbit and offspring from founder lines 1, 2 and 3. The offspring from founder lines 2 and 3 showed higher levels of HLA-A2.1 protein on their PBLs than the offspring from founder line 1. No positive signals were found in normal control rabbit PBLs.
FIG. 3.
Expression of rabbit MHC-I on cell surfaces of PBLs from offspring from different founder rabbit lines. The offspring of founder rabbit line 1 (A) showed slightly higher levels of MHC-I than their nontransgenic siblings, while opposite results were found in founder rabbit lines 2 and 3 (B). However, these differences were not statistically significant (unpaired Student's t test; P > 0.05). MFI, mean fluorescence intensity.
FIG. 4.
HLA-A2.1 and MHC-I expression levels in different tissues. (A) Biopsy samples collected from transgenic rabbits' ears were frozen in liquid nitrogen and stored at −70°C. The frozen slices of ear tissues were stained with anti-HLA-A2.1 (BB7.2) and anti-rabbit MHC-I monoclonal antibodies by standard immunohistochemistry. The HLA-A2.1 and rabbit MHC-I expression levels shared similar patterns in different generations (F1 to F6) of HLA-A2.1 transgenic rabbits. The HLA-A2.1 expression levels were stable in the offspring of all the founder rabbits. (B) Tongue, spleen, and back-skin tissues from a transgenic rabbit were labeled with anti-HLA-A2.1 and anti rabbit MHC-I monoclonal antibodies. Both proteins show similar expression patterns in all the tissues; the highest expression levels were found in the spleen tissue.
We also tested the distribution of HLA-A2.1 and rabbit MHC-I in different organs. To do this, we sacrificed one transgenic rabbit and took biopsy samples from the spleen, liver, lungs, tongue, ear, and back skin. HLA-A2.1 and rabbit MHC-I were detected in all tissues tested, with the highest levels detected in spleen tissues (Fig. 4B).
CRPV DNA infection in HLA-A2.1 transgenic rabbits.
In a previous study, we demonstrated that HLA-A2.1 transgenic rabbit cells could process and present an HLA-A2.1-specific epitope (19). We then wanted to test if these rabbits showed comparable immunity to CRPV infection. Seven HLA-A2.1 transgenic (rabbits 2990, 2992, and 2994 were offspring of founder line 1 bred with an outbred rabbit, while rabbits 602, 603, 604, and 606 were offspring of founder line 1 bred with an inbred EIII/JC rabbit) and two nontransgenic rabbits (rabbits 2987 and 601 were siblings of these respective transgenic rabbits) were challenged with two Hershey CRPV strains (H.CRPVp is a progressive strain and H.CRPVr is a regressive strain) on left and right back skin sites, respectively (16). H.CRPVp-induced papillomas persisted on four of the seven transgenic rabbits and two normal rabbits, while H.CRPVr-induced papillomas regressed on all the rabbits (Table 1). Interestingly, H.CRPVp-induced papillomas on three of the seven HLA-A2.1 rabbits regressed, although the rabbit MHC-I protein levels in these rabbits were comparable to those in control normal rabbits (data not shown).
TABLE 1.
Papilloma evolution in HLA-A2.1 transgenic rabbits after wild-type progressive (H.CRPVp) and regressive (H.CRPVr) CRPV DNA challenge
| Genetic background and rabbit | Expression levels ofa:
|
Papilloma evolution withb:
|
||
|---|---|---|---|---|
| HLA-A2.1 | Rabbit MHC-I | H.CRPVp | H.CRPVr | |
| Outbred NZW | ||||
| 2987 | − | +++ | P | R |
| 2990 | ++ | ++ | P | R |
| 2992 | ++ | ++ | P | R |
| 2994 | ++ | ++ | R | R |
| Outbred NZW and inbred EIII/JC | ||||
| 601 | − | ++ | P | R |
| 602 | +++ | +++ | R | R |
| 603 | +++ | +++ | R | R |
| 604 | ++ | ++ | P | R |
| 607 | ++ | ++ | P | R |
Subjective scales for the expression levels of A2 and MHC-I were used. −, negative signal; +, positive signal, with ++ and +++ representing higher and the highest levels of positive staining.
P, papilloma persisted; R, papilloma regressed.
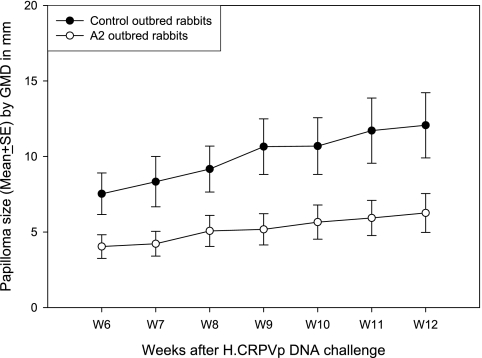
In a more-controlled experiment, we checked papilloma outgrowth induced by H.CRPVp DNA infection in both normal and HLA-A2.1 transgenic rabbits (Table 2). The HLA-A2.1 transgenic rabbits grew significantly smaller papillomas than the nontransgenic rabbits (unpaired Student's t test; P < 0.05) (Fig. 5). Significantly more H.CRPVp-induced papillomas regressed in the HLA-A2.1 transgenic rabbits than in the nontransgenic rabbits, and this was also true in rabbits that were used as ubiquitin-vaccinated control animals from previous epitope DNA vaccination experiments (Table 2) (Fisher's exact test; P < 0.05) (18). These findings suggested that the HLA-A2.1 transgenic rabbits might target some naturally existing HLA-A2.1-restricted epitopes from CRPV proteins more effectively than native rabbit MHC-I proteins.
TABLE 2.
Papilloma regression rates in HLA-A2.1 transgenic and normal rabbits after H.CRPVp infection
| Treatment group | Rabbit genotype | No. of challenged animals (no. of sites) | No. of animals with regression (no. of regressed sites) | % Regression rate of animals (% sites)b |
|---|---|---|---|---|
| Ubiquitin vaccinea | HLA-A2.1 | 11 (44) | 1 (10) | 9 (22.7*) |
| Normal | 7 (28) | 0 (1) | 0 (3.6) | |
| No vaccine | HLA-A2.1 | 11 (44) | 4 (18) | 36 (41*) |
| Normal | 6 (24) | 0 (0) | 0 (0) |
Ubiquitin vaccine was a vector control for previous studies (19).
*, P < 0.05 compared to normal rabbits, by Fisher's exact test.
FIG. 5.
Papilloma outgrowth after H.CRPVp DNA infection in both HLA-A2.1 and normal rabbits. Significantly smaller papillomas were found in HLA-A2.1 transgenic rabbits than in normal rabbits (unpaired Student's t test; P < 0.05).
L1 expression and anti-L1 antibody generation in HLA-A2.1 transgenic rabbits.
We have demonstrated that CRPV virus infection generates anti-L1 antibody in domestic rabbits and that abundant L1 can be detected in situ, implying the presence of virions (data not shown). Here, we also tested anti-L1 antibody generation and virus production in HLA-A2.1 transgenic rabbits from both outbred and inbred EIII/JC genetic backgrounds. Sera collected from these rabbits were tested in a routine ELISA. Our data showed that all CRPV- and three out of five DNA-challenged rabbits generated high titers of anti-L1 antibody (Table 3). Using standard immunohistochemistry tests, we also detected L1 protein in CRPV-induced papillomas in these transgenic rabbits (Fig. 6). Therefore, the HLA-A2.1 transgenic rabbits showed an infection pattern comparable to that of normal rabbits. All extracts from CRPV-induced papillomas were able to produce E1^E4 transcripts, as shown by the results of an in vitro infectivity assay (unpublished data).
TABLE 3.
L1 protein expression and anti-CRPV L1 antibody generation after infection with H.CRPVp virus and/or DNA challenge in HLA-A2.1 transgenic rabbits
| Type of challenge | Rabbit | L1 expression levela | Anti-L1 antibody level (OD value) | Titer |
|---|---|---|---|---|
| Infectious virus (1:100) | 811 | NA | 0.336 | 12,800 |
| 812 | ++++ | 0.254 | 25,600 | |
| 813 | +++ | 0.436 | 12,800 | |
| 814 | ND | 0.540 | 12,800 | |
| 815 | ++++ | 0.656 | >25,600 | |
| 816 | ++ | 0.255 | 12,800 | |
| 817 | NA | 0.429 | 6,400 | |
| 818 | + | 0.377 | 12,800 | |
| 819 | ++/+++ | 0.307 | >25,600 | |
| 820 | ++ | 0.388 | 12,800 | |
| 821 | NA | 0.498 | 10,000 | |
| CRPVp DNA (10 μg) | 802 | + | 0.398 | 12,800 |
| 805 | + | 0.386 | 12,800 | |
| 808 | + | 0.374 | 12,800 | |
| 837 | + | 0.009 | ND | |
| 849 | + | 0.012 | ND |
NA, no papillomas available for histological analysis; ND, not done; +, positive signal, with ++, +++, and ++++ representing higher and the highest levels of positive signals.
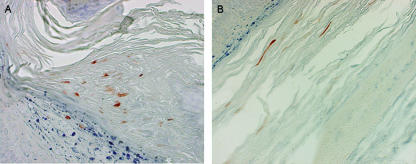
FIG. 6.
L1 protein expression in H.CRPVp-induced papillomas. Biopsy samples harvested from H.CRPVp- and viral DNA-induced papillomas from HLA-A2.1 transgenic rabbits were labeled with a specific anti-CRPV L1 monoclonal antibody. High and low levels of L1 were detected in virus-induced (A) and viral DNA-induced (B) papilloma tissues, respectively.
Cancer development in CRPV DNA-induced papillomas.
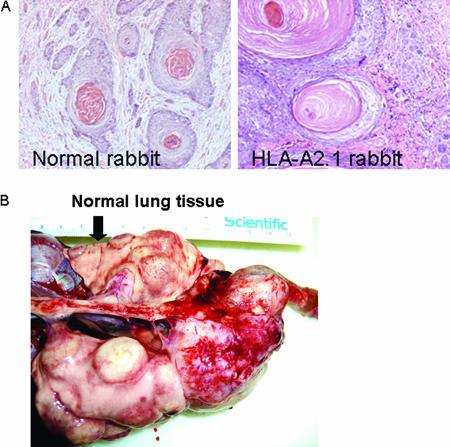
Although we did not track cancer development in most HLA-A2.1 transgenic rabbits because of increased immunity in these animals, H.CRPVp-induced papillomas were capable of malignant progression in HLA-A2.1 transgenic rabbits. Two out of five H.CRPVp challenge sites on one HLA-A2.1 transgenic rabbit developed cancer around 13 months after infection. Another HLA-A2.1 transgenic rabbit developed metastatic cancer in the lungs (Fig. 7B) at 24 months after infection. Histological examination showed typical cancer morphology in these malignant tissues when compared to those cancers developing in nontransgenic rabbits (Fig. 7A).
FIG. 7.
(A) Hematoxylin and eosin histological staining of malignant tumor tissues induced by infection with H.CRPVp DNA in normal and HLA-A2.1 transgenic rabbits; (B) H.CRPVp DNA-induced metastatic cancer in lung tissue of one HLA-A2.1 transgenic rabbit.
DISCUSSION
We report here a novel HLA-A2.1 transgenic rabbit model and the results showing their responses to experimental CRPV infections. Three transgenic founder lines, while showing different sizes of the transgene by Southern blot analysis, all expressed HLA-A2.1 on cell surfaces within different organs. We assume that the larger size (about twofold the expected size) of the transgene in founder line 1 reflects duplications and the loss of an EcoRI site upon integration in the genome. However, this change did not disturb the expression of HLA-A2.1 protein in this rabbit. The offspring from these founder rabbits maintained HLA-A2.1 transgene expression. HLA-A2.1 colocalized with rabbit MHC-I on cell surfaces and did not appear to diminish concurrent rabbit MHC-I expression (19). HLA-A2.1 transgenic rabbits were susceptible to CRPV infection similarly to normal rabbits but with higher regression rates. The higher HLA-A2.1 levels correlated with stronger natural immune responses to CRPV infection, leading to increased rates of papilloma regression in some animals. Our previous studies demonstrated that higher spontaneous regression rates were found among inbred EIII/JC rabbits than in normal outbred NZW rabbits (16). In this study, we also found that HLA-A2.1 transgenic rabbits containing a more-inbred EIII/JC genetic background showed higher regression rates than rabbits containing a less inbred background (Table 1 and 2). A malignant progression of some tumors was also found in HLA-A2.1 transgenic rabbits. Therefore, HLA-A2.1 integrated into the rabbit genome is compatible with rabbit immune system function.
HLA-A2.1 transgenic mice have been used extensively to screen and test HLA-A2.1-restricted epitopes for the development of vaccines for stimulating effective and specific host immunities (14, 20, 23, 30). HPVs are highly species-specific tumor viruses, and no rodent papillomavirus infection model is available to study papillomavirus infection of rodents in vivo (4). Rabbits, together with cattle and domestic dogs, are animal models widely used for studying virus-host interactions (3). CRPV infection mimics high-risk HPV-induced cancer in humans and is therefore advantageous for studying malignancies initiated by papillomavirus infections (5). An additional advantage of the rabbit system is that the CRPV genome has a large capacity for modification, which allows investigators to generate hybrid papillomavirus genomes and to test their immunogenicities and oncogenicities in vivo (15). The HLA-A2.1 transgenic rabbits are thus useful animal models for the development and testing of therapeutic vaccines against papillomavirus infections.
In addition to its potential application to HPV vaccine development, the HLA-A2.1 transgenic rabbit model can also be used to study other human pathogens to which rabbits are susceptible, such as human T-cell leukemia virus types I and II (10), Epstein-Barr virus-like viruses (21), adenovirus type 5 (12), and herpes simplex virus type 1 (26) and nonviral pathogens such as those causing tuberculosis (6) and syphilis (9). These infectious agents have shown pathogenesis in rabbits that is comparable to that found in human infections. We can screen HLA-A2.1-restricted epitopes from these pathogens and test their immunogenicities in rabbits, as has been initiated for papillomavirus epitopes. We believe that the HLA-A2.1 transgenic rabbit model shows promise for investigations in the fields of immunology, vaccine development, and virology.
Acknowledgments
We thank Martin Pickel for excellent help with the animals.
This work was supported by National Cancer Institute grant R01 CA47622 from the National Institutes of Health and by the Jake Gittlen Memorial Golf Tournament.
Footnotes
Published ahead of print on 25 April 2007.
REFERENCES
- 1.Brandsma, J. L. 2005. The cottontail rabbit papillomavirus model of high-risk HPV-induced disease. Methods Mol. Med. 119:217-235. [DOI] [PubMed] [Google Scholar]
- 2.Brandsma, J. L., and W. Xiao. 1993. Infectious virus replication in papillomas induced by molecularly cloned cottontail rabbit papillomavirus DNA. J. Virol. 67:567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campo, M. S. 2002. Animal models of papillomavirus pathogenesis. Virus Res. 89:249-261. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, N. D. 2005. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir. Chem. Chemother. 16:355-362. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, N. D., R. Han, and J. W. Kreider. 2000. Cottontail rabbit papillomavirus (CRPV), p. 485-502. In R. Ahmed and I. Chen (ed.), Persistent viral infections. John Wiley & Sons Ltd., Chichester, England.
- 6.Dorman, S. E., C. L. Hatem, S. Tyagi, K. Aird, J. Lopez-Molina, M. L. Pitt, B. C. Zook, A. M. Dannenberg, Jr., W. R. Bishai, and Y. C. Manabe. 2004. Susceptibility to tuberculosis: clues from studies with inbred and outbred New Zealand White rabbits. Infect. Immun. 72:1700-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, T. G., W. Bonnez, R. C. Rose, S. Koenig, L. Demeter, J. A. Suzich, D. O'Brien, M. Campbell, W. I. White, J. Balsley, and R. C. Reichman. 2001. A phase 1 study of a recombinant viruslike particle vaccine against human papillomavirus type 11 in healthy adult volunteers. J. Infect. Dis. 183:1485-1493. [DOI] [PubMed] [Google Scholar]
- 8.Fausch, S. C., D. M. Da Silva, G. L. Eiben, I. C. Le Poole, and W. M. Kast. 2003. HPV protein/peptide vaccines: from animal models to clinical trials. Front. Biosci. 8:S81-S91. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald, T. J., and M. K. Froberg. 1991. Congenital syphilis in newborn rabbits: immune functions and susceptibility to challenge infection at 2 and 5 weeks of age. Infect. Immun. 59:1869-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchini, G., J. Tartaglia, P. Markham, J. Benson, J. Fullen, M. Wills, J. Arp, G. Dekaban, E. Paoletti, and R. C. Gallo. 1995. Highly attenuated HTLV type I-env poxvirus vaccines induce protection against a cell-associated HTLV type I challenge in rabbits. AIDS Res. Hum. Retrovir. 11:307. [DOI] [PubMed] [Google Scholar]
- 11.Frazer, I. H. 2004. Prevention of cervical cancer through papillomavirus vaccination. Nat. Rev. Immunol. 4:46-54. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, Y. J., E. Romanowski, and T. Araullo-Cruz. 1992. An ocular model of adenovirus type 5 infection in the NZ rabbit. Investig. Ophthalmol. Vis. Sci. 33:574-580. [PubMed] [Google Scholar]
- 13.Han, R., N. M. Cladel, C. A. Reed, X. Peng, and N. D. Christensen. 1999. Protection of rabbits from viral challenge by gene gun-based intracutaneous vaccination with a combination of cottontail rabbit papillomavirus E1, E2, E6, and E7 genes. J. Virol. 73:7039-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himoudi, N., J. D. Abraham, A. Fournillier, Y. C. Lone, A. Joubert, D. B. Op, D. Freida, F. Lemonnier, M. P. Kieny, and G. Inchauspe. 2002. Comparative vaccine studies in HLA-A2.1-transgenic mice reveal a clustered organization of epitopes presented in hepatitis C virus natural infection. J. Virol. 76:12735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, J., N. M. Cladel, K. Balogh, L. Budgeon, and N. D. Christensen. 2007. Impact of genetic changes to the CRPV genome and their application to the study of pathogenesis in vivo. Virology. 358:384-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, J., N. M. Cladel, M. D. Pickel, and N. D. Christensen. 2002. Amino acid residues in the carboxy-terminal region of cottontail rabbit papillomavirus E6 influence spontaneous regression of cutaneous papillomas. J. Virol. 76:11801-11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J., R. Han, N. M. Cladel, M. D. Pickel, and N. D. Christensen. 2002. Intracutaneous DNA vaccination with the E8 gene of cottontail rabbit papillomavirus induces protective immunity against virus challenge in rabbits. J. Virol. 76:6453-6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, J., X. Peng, N. M. Cladel, M. D. Pickel, and N. D. Christensen. 2005. Large cutaneous rabbit papillomas that persist during cyclosporin A treatment can regress spontaneously after cessation of immunosuppression. J. Gen. Virol. 86:55-63. [DOI] [PubMed] [Google Scholar]
- 19.Hu, J., X. Peng, T. D. Schell, L. R. Budgeon, N. M. Cladel, and N. D. Christensen. 2006. An HLA-A2.1-transgenic rabbit model to study immunity to papillomavirus infection. J. Immunol. 177:8037-8045. [DOI] [PubMed] [Google Scholar]
- 20.Kawakami, Y., R. Zakut, S. L. Topalian, H. Stötter, and S. A. Rosenberg. 1992. Shared human melanoma antigens: recognition by tumor-infiltrating lymphocytes in HLA-A2.1-transfected melanomas. J. Immunol. 148:638-643. [PubMed] [Google Scholar]
- 21.Koirala, T. R., K. Hayashi, Z. Jin, S. Onoda, T. Tanaka, W. Oda, K. Ichimura, N. Ohara, T. Oka, M. Yamada, and T. Yoshino. 2004. Induction and prevention of virus-associated malignant lymphoma by serial transmission of EBV-related virus from cynomolgus by blood transfusion in rabbits. Acta Med. Okayama 58:67-74. [DOI] [PubMed] [Google Scholar]
- 22.Kreider, J. W., N. M. Cladel, S. D. Patrick, P. A. Welsh, S. L. DiAngelo, J. M. Bower, and N. D. Christensen. 1995. High efficiency induction of papillomas in vivo using recombinant cottontail rabbit papillomavirus DNA. J. Virol. Methods 55:233-244. [DOI] [PubMed] [Google Scholar]
- 23.Le, A. X., E. J. Bernhard, M. J. Holterman, S. Strub, P. Parham, E. Lacy, and V. H. Engelhard. 1989. Cytotoxic T cell responses in HLA-A2.1 transgenic mice. Recognition of HLA alloantigens and utilization of HLA-A2.1 as a restriction element. J. Immunol. 142:1366-1371. [PubMed] [Google Scholar]
- 24.Leachman, S. A., M. Shylankevich, M. D. Slade, D. Levine, R. K. Sundaram, W. Xiao, M. Bryan, D. Zelterman, R. E. Tiegelaar, and J. L. Brandsma. 2002. Ubiquitin-fused and/or multiple early genes from cottontail rabbit papillomavirus as DNA vaccines. J. Virol. 76:7616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowy, D. R., and J. T. Schiller. 2006. Prophylactic human papillomavirus vaccines. J. Clin. Investig. 116:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majumdar, S., Y. E. Nashed, K. Patel, R. Jain, M. Itahashi, D. M. Neumann, J. M. Hill, and A. K. Mitra. 2005. Dipeptide monoester ganciclovir prodrugs for treating HSV-1-induced corneal epithelial and stromal keratitis: in vitro and in vivo evaluations. J. Ocul. Pharmacol. Ther. 21:463-474. [DOI] [PubMed] [Google Scholar]
- 27.Nicholls, P. K., and M. A. Stanley. 2000. The immunology of animal papillomaviruses. Vet. Immunol. Immunopathol. 73:101-127. [DOI] [PubMed] [Google Scholar]
- 28.Peng, X., R. O. Olson, C. B. Christian, C. M. Lang, and J. W. Kreider. 1993. Papillomas and carcinomas in transgenic rabbits carrying EJ-ras DNA and cottontail rabbit papillomavirus DNA. J. Virol. 67:1698-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poland, G. A., R. M. Jacobson, L. A. Koutsky, G. M. Tamms, R. Railkar, J. F. Smith, J. T. Bryan, P. F. Cavanaugh, Jr., K. U. Jansen, and E. Barr. 2005. Immunogenicity and reactogenicity of a novel vaccine for human papillomavirus 16: a 2-year randomized controlled clinical trial. Mayo Clin. Proc. 80:601-610. [DOI] [PubMed] [Google Scholar]
- 30.Schell, T. D., J. D. Lippolis, and S. S. Tevethia. 2001. Cytotoxic T lymphocytes from HLA-A2.1 transgenic mice define a potential human epitope from simian virus 40 large T antigen. Cancer Res. 61:873-879. [PubMed] [Google Scholar]
- 31.Stanley, M. A. 2003. Progress in prophylactic and therapeutic vaccines for human papillomavirus infection 3. Expert Rev. Vaccines 2:381-389. [DOI] [PubMed] [Google Scholar]
- 32.Villa, L. L., R. L. Costa, C. A. Petta, R. P. Andrade, K. A. Ault, A. R. Giuliano, C. M. Wheeler, L. A. Koutsky, C. Malm, M. Lehtinen, F. E. Skjeldestad, S. E. Olsson, M. Steinwall, D. R. Brown, R. J. Kurman, B. M. Ronnett, M. H. Stoler, A. Ferenczy, D. M. Harper, G. M. Tamms, J. Yu, L. Lupinacci, R. Railkar, F. J. Taddeo, K. U. Jansen, M. T. Esser, H. L. Sings, A. J. Saah, and E. Barr. 2005. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol. 6:271-278. [DOI] [PubMed] [Google Scholar]