Abstract
The susceptibility of sheep to classical scrapie and bovine spongiform encephalopathy (BSE) is mainly influenced by prion protein (PrP) polymorphisms A136V, R154H, and Q171R, with the ARR allele associated with significantly decreased susceptibility. Here we report the protective effect of the amino acid substitution M137T, I142K, or N176K on the ARQ allele in sheep experimentally challenged with either scrapie or BSE. Such observations suggest the existence of additional PrP alleles that significantly decrease the susceptibility of sheep to transmissible spongiform encephalopathies, which may have important implications for disease eradication strategies.
Scrapie is a fatal and infectious disease of sheep and goats belonging to a group of neurodegenerative disorders affecting humans and animals, known as transmissible spongiform encephalopathies (TSEs) or prion diseases. Susceptibility to TSEs is influenced by both the genotype, mainly represented by the prion protein (PrP) encoded by the host, and by the TSE strain involved.
The ovine PrP gene is highly polymorphic, and new variants have been described in the last years, bringing the total number of known amino acid sequences to more than 45. The most common polymorphisms at codons 136, 154, and 171, generating the ARR (alanine at codon 136 and arginine at codons 154 and 171), ARH, AHQ, ARQ, and VRQ alleles, have been associated with different levels of susceptibility to the disease (3, 4, 6, 13, 17). Only these five alleles have been tested frequently by experimental challenges of sheep with classical scrapie, and the ARR allele was always found to be associated with partial or total protection from disease (16). On this basis, selective sheep breeding programs aimed at increasing the frequency of the ARR allele have been implemented in Europe and the United States. However, the recent findings that ARR/ARR sheep are susceptible after intracerebral (i.c.) challenge with bovine spongiform encephalopathy (BSE) (15) and may be affected by atypical scrapie (8) question the reliance on the ARR allele for eradication strategies. The study of the association of rare PrP genotypes with scrapie susceptibility would be of paramount importance for eradication strategies, but it is hindered by the low frequencies of most of these PrP variants. To date, only one study has reported an experimental challenge of sheep carrying a rare PrP allele, and it demonstrated that sheep heterozygous for the ARL168Q variant have increased resistance to BSE (12).
Evidence is reported here on the protective effects of M137T, I142K, and N176K polymorphisms in sheep experimentally challenged with either scrapie or BSE.
Ewes from a single flock of the Sarda breed with no history of scrapie were initially genotyped at codons 136, 154, and 171 by using a rapid genotyping method (22). One hundred animals carrying genotypes representative of the Sarda breed (2, 24) were selected and experimentally challenged with scrapie or BSE. The age of animals at the time of inoculation ranged from 7 to 8 and 9 to 10 months for orally and i.c. scrapie-challenged animals, respectively, and from 8 to 9 months for i.c. BSE-challenged sheep.
The scrapie inoculum was prepared by pooling brain tissues from eight fully sequenced ARQ/ARQ scrapie-affected Sarda sheep, while the BSE one was prepared by pooling brain portions from two affected cows (TSE Archive, United Kingdom). The ewes were challenged either i.c. (1 ml of 10% brain homogenate) or by the oral route (15 ml of 30% brain homogenate), treated according to European Union recommendations for animal welfare, and housed in high-confinement facilities. Euthanasia was performed when clinical signs consistent with TSE were clearly present. Diagnosis of TSE was confirmed by detection of the disease-specific PrP (PrPSc) at the obex by Western blotting (23).
Observations on the effects of rare PrP polymorphisms on the scrapie susceptibility of goats (1, 23) as well as sheep affected by the Nor98 strain (18) prompted us to determine the entire PrP coding sequence for sheep challenged with scrapie and BSE, as previously described (24).
The sequence analysis revealed the presence of the M137T (n = 12), N176K (n = 7), and I142K (n = 1) allelic variants of the ARQ allele. The first two of these variants have already been described (6, 21). The AK142RQ allele is reported here for the first time (with a T-to-A transversion in the second position of codon 142), although a different polymorphism at the same codon has already been described (25). Table 1 summarizes the updated PrP genotypes, which were reconstructed on the assumption that all polymorphisms are mutually exclusive.
TABLE 1.
PrP genotypes and survival times for sheep challenged with scrapie (i.c. or orally) or BSE (i.c.)
| PrP genotype and disease challenge | No. of affected sheep/no. of inoculated sheep | Mean survival time ± SD (days) |
|---|---|---|
| i.c. scrapie transmission | ||
| ARQ/ARQ | 5/5 | 462 ± 25 |
| ARQ/AHQ | 5/5 | 703 ± 36 |
| AHQ/AHQ | 1/1 | 790 |
| ARQ/ARH | 1/1 | 1,083 |
| AHQ/ARH | 3/3 | 1,252 ± 87 |
| ARQ/ARR | 0/13 | >1,450 |
| AHQ/ARR | 0/1 | >1,450 |
| ARH/ARR | 0/1 | >1,450 |
| ARR/ARR | 0/5 | >1,450 |
| AT137RQ/ARQK176 | 0/1 | >1,450 |
| ARQK176/AHQ | 0/1 | >1,450 |
| ARQK176/ARR | 0/2 | >1,450 |
| Oral scrapie transmission | ||
| ARQ/ARQ | 5/5 | 860 ± 16 |
| ARQ/AHQ | 5/5 | 1,115 ± 159 |
| ARQ/ARH | 0/3 | >1,550 |
| ARQ/ARR | 0/10 | >1,550 |
| ARH/ARR | 0/1 | >1,550 |
| ARR/ARR | 0/5 | >1,550 |
| ARQ/AT137RQ | 0/3 | >1,550 |
| ARQ/ARQK176 | 0/1 | >1,550 |
| ARQ/AK142RQ | 0/1 | >1,550 |
| ARQK176/ARR | 0/1 | >1,550 |
| i.c. BSE transmission | ||
| ARQ/ARQ | 2/2 | 480 ± 2 |
| ARQ/ARH | 2/2 | 748 ± 18 |
| ARQ/ARR | 0/5 | >950 |
| AHQ/ARR | 0/1 | >950 |
| ARR/ARR | 0/7 | >950 |
| ARQ/AT137RQ | 0/3 | >950 |
| ARQK176/AHQ | 0/1 | >950 |
| AT137RQ/AHQ | 0/1 | >950 |
| AT137RQ/ARH | 0/1 | >950 |
| AT137RQ/ARR | 0/3 | >950 |
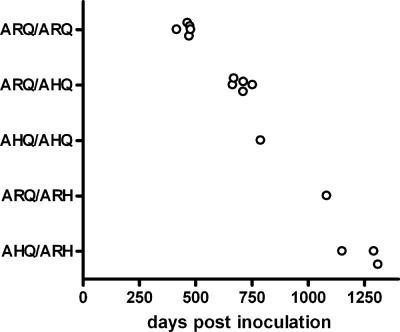
After i.c. scrapie challenge, all sheep with the ARQ/ARQ (n = 5), ARQ/AHQ (n = 5), AHQ/AHQ (n = 1), ARQ/ARH (n = 1), and AHQ/ARH (n = 3) genotypes succumbed to the disease, with mean survival times ranging from 462 days postinfection (dpi) for ARQ/ARQ sheep to 1,252 dpi for AHQ/ARH sheep. The diverse survival times observed (Fig. 1) indicate that these PrP alleles are associated with different levels of susceptibility to scrapie, with a rank order as follows: ARQ > AHQ > ARH. Similarly, after oral scrapie challenge, sheep with the ARQ/ARQ (n = 5) and ARQ/AHQ (n = 5) genotypes showed terminal disease at 860 ± 16 and 1,115 ± 159 dpi, respectively, while ARQ/ARH sheep (n = 3) were healthy at the time of writing of this report.
FIG. 1.
Scatter plot of survival times of individual sheep with ARQ/ARQ, ARQ/AHQ, AHQ/AHQ, ARQ/ARH, and AHQ/ARH genotypes after i.c. challenge with scrapie.
In clear contrast, sheep carrying the M137T (n = 4), N176K (n = 3), and I142K (n = 1) variations in the ARQ/ARQ and ARQ/AHQ backgrounds did not show any signs of disease (up to 1,450 and 1,550 dpi after i.c. and oral challenges, respectively). Animals with the ARQ/ARR (n = 23), ARH/ARR (n = 2), AHQ/ARR (n = 1), ARQK176/ARR (n = 3), and ARR/ARR (n = 10) genotypes were also healthy at the time of this report.
Interestingly, in a parallel experiment which will be reported separately, aimed at studying the kinetics of PrPSc accumulation in tissues of Sarda sheep orally infected with the same scrapie inoculum and dose as those used in the present experiment, corroborating findings were observed. Single sheep with the ARQ/ARQK176 and ARQ/AK142RQ genotypes, culled at 9 and 12 months postinfection (p.i.), respectively, were PrPSc negative in the lymph reticular system (LRS) as well as in the central nervous system (CNS), whereas all ARQ/ARQ animals culled at the same intervals were already positive at the LRS level. Furthermore, one ARQ/AT137RQ sheep euthanized at 20 months p.i. was still negative in both the LRS and CNS, while all ARQ/ARQ sheep showed PrPSc deposition in both the LRS and CNS from 16 months p.i. onwards.
After i.c. challenge with BSE, all sheep with the ARQ/ARQ (n = 2) and ARQ/ARH (n = 2) genotypes succumbed to the disease, with survival times of 480 ± 2 and 748 ± 18, respectively. Conversely, sheep carrying the M137T (n = 5) or N176K (n = 1) variation in the ARQ/ARQ, ARQ/AHQ, or ARQ/ARH background did not show any signs of the disease (up to 950 dpi). Sheep with the ARQ/ARR (n = 5), AHQ/ARR (n = 1), AT137RQ/ARR (n = 3), or ARR/ARR (n = 7) genotype were also healthy at the time of this report.
The AT137RQ and ARQK176 alleles have never been reported for diseased sheep from scrapie-affected flocks, but their frequencies in healthy controls were too low to evidence any significant protective effect (6, 21, 24). In Italy, since 2004, we have officially subjected all confirmed scrapie cases to PrP genotype determination by sequencing analysis. Of 187 ARQ/ARQ sheep with classical scrapie, only 15 carried additional polymorphisms, namely, L141F (n = 10), M112T (n = 4), and G127V/L141F (n = 1). Interestingly, no variation was found at codon 137, 142, or 176. The frequencies of polymorphisms at codons 137, 142, and 176 in the Italian sheep population are unknown, thus preventing any statistical analysis. However, in two examined flocks of the Sarda breed (n = 344), the frequencies of these polymorphisms were 7%, 1%, and 2%, respectively. Furthermore, data from the literature suggest that the AT137RQ allele might be quite common in sheep. Indeed, it has been observed in several breeds and countries (6, 9, 14, 20, 21, 24) and was found in 19.4% of flocks examined in the United Kingdom (11).
Several lines of evidence support the conclusion that the AT137RQ, AK142RQ, and ARQK176 alleles are associated with significantly decreased susceptibility to scrapie. Indeed, sheep with these alleles (i) were still healthy 1,450 days after i.c. challenge, while all sheep with susceptible genotypes succumbed to the disease; (ii) were still healthy 1,550 days after oral challenge and, when culled at different intervals p.i., were negative for PrPSc in the LRS and CNS; and (iii) were not found among any sheep with classical scrapie reported in Italy since 2004. To our knowledge, this is the first report of PrP alleles other than the ARR allele which are able to confer a protective effect after experimental challenge with scrapie. A possible limitation of our study is that the mutated alleles were only challenged in heterozygosity. Indeed, it has been reported for mice inoculated with some scrapie strains that heterozygous individuals could be less susceptible than homozygous individuals (7). This phenomenon, known as overdominance of heterozygosis (10), has never been reported so far for sheep scrapie. For instance, the ARR allele has been shown to be either partially or completely dominant (16). In the present study, heterozygous R154H or Q171H sheep and double heterozygous R154H/Q171H sheep were fully susceptible to i.c. challenge with scrapie or BSE, suggesting that the protection observed in sheep carrying the mutated alleles is not due to heterozygosis per se. However, interim results from ongoing UK studies with BSE suggest that the ARR allele, although associated with clearly decreased susceptibility, could be overdominant, conferring more protection to ARQ/ARR than to ARR/ARR sheep (15). In this respect, further experiments are needed to study the susceptibility of sheep carrying the mutated alleles in homozygosis.
We do not know if the observed protective effect could be extended to other TSE strains. The AT137RQ allele was previously analyzed by in vitro conversion experiments and was reported to be inconvertible to PrPSc with a VRQ scrapie isolate but partially convertible with an ARQ isolate (5). Although in vitro conversion experiments may not always mirror in vivo experimental results (19), these findings could suggest that the behavior of the AT137RQ allele may vary with different scrapie isolates. However, the observation that all animals with the AT137RQ and ARQK176 alleles were still healthy 1,550 and 950 days, respectively, after challenge with high doses of either scrapie or BSE is impressive and could have important implications for disease eradication strategies. Nevertheless, it is too early to state if these polymorphisms will confer full protection from BSE, since only 950 days have elapsed from the challenge. The challenged sheep will be maintained alive to verify if animals will develop the disease in the future.
We showed that the low-frequency PrP variants described in the present study are associated with a high level of protection from scrapie and confer at least partial protection after i.c. BSE challenge. Taking into account the recently reported increased resistance effect of the ARL168Q allele in response to BSE (12), it could be envisaged that other alleles having a protective effect could exist in sheep and that selection programs for different resistance alleles could ensure better protection against TSEs in view of the possible circulation of BSE in ovine populations or the appearance of new strains targeting different sheep genotypes.
Nucleotide sequence accession number.
The sequence of the AK142RQ variant is available under GenBank accession number EF153678.
Acknowledgments
We thank the veterinary laboratory network of Istituti Zooprofilattici Sperimentali for its efficient activity in the frame of scrapie surveillance. We also thank Maurice Bardsley from the TSE Archive (United Kingdom) for kindly supplying BSE tissues and Consiglia Parisi and Nadia Palazzini for their skillful assistance.
This work was supported by two grants from the Italian Ministry of Health (Progetti Strategici “Encefalopatie spongiformi trasmissibili dell'uomo e degli animali,” ricerca finalizzata 1% anni 2001 e 2002).
Footnotes
Published ahead of print on 18 April 2007.
REFERENCES
- 1.Acutis, P. L., A. Bossers, J. Priem, M. V. Riina, S. Peletto, M. Mazza, C. Casalone, G. Forloni, G. Ru, and M. Caramelli. 2006. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. J. Gen. Virol. 87:1029-1033. [DOI] [PubMed] [Google Scholar]
- 2.Agrimi, U., M. Conte, L. Morelli, M. A. Di Bari, G. Di Guardo, C. Ligios, G. Antonucci, G. M. Aufiero, N. Pozzato, F. Mutinelli, R. Nonno, and G. Vaccari. 2003. Animal transmissible spongiform encephalopathies and genetics. Vet. Res. Commun. 27(Suppl. 1):31-38. [DOI] [PubMed] [Google Scholar]
- 3.Baylis, M., C. Chihota, E. Stevenson, W. Goldmann, A. Smith, K. Sivam, S. Tongue, and M. B. Gravenor. 2004. Risk of scrapie in British sheep of different prion protein genotype. J. Gen. Virol. 85:2735-2740. [DOI] [PubMed] [Google Scholar]
- 4.Belt, P. B., I. H. Muileman, B. E. Schreuder, J. Bos-de Ruijter, A. L. Gielkens, and M. A. Smits. 1995. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J. Gen. Virol. 76:509-517. [DOI] [PubMed] [Google Scholar]
- 5.Bossers, A., R. de Vries, and M. A. Smits. 2000. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J. Virol. 74:1407-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossers, A., B. E. Schreuder, I. H. Muileman, P. B. Belt, and M. A. Smits. 1996. PrP genotype contributes to determining survival times of sheep with natural scrapie. J. Gen. Virol. 77:2669-2673. [DOI] [PubMed] [Google Scholar]
- 7.Bruce, M. E. 1993. Scrapie strain variation and mutation. Br. Med. Bull. 49:822-838. [DOI] [PubMed] [Google Scholar]
- 8.Buschmann, A., G. Luhken, J. Schultz, G. Erhardt, and M. H. Groschup. 2004. Neuronal accumulation of abnormal prion protein in sheep carrying a scrapie-resistant genotype (PrPARR/ARR). J. Gen. Virol. 85:2727-2733. [DOI] [PubMed] [Google Scholar]
- 9.DeSilva, U., X. Guo, D. M. Kupfer, S. C. Fernando, A. T. Pillai, F. Z. Najar, S. So, G. Q. Fitch, and B. A. Roe. 2003. Allelic variants of ovine prion protein gene (PRNP) in Oklahoma sheep. Cytogenet. Genome Res. 102:89-94. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson, A. G., and H. Fraser. 1979. An assessment of the genetics of scrapie in sheep and mice, p. 367-385. In S. B. Prusiner and W. J. Hadlow (ed.), Slow transmissible diseases of the nervous system, vol. 1. Academic Press, New York, NY. [Google Scholar]
- 11.Goldmann, W., M. Baylis, C. Chihota, E. Stevenson, and N. Hunter. 2005. Frequencies of PrP gene haplotypes in British sheep flocks and the implications for breeding programmes. J. Appl. Microbiol. 98:1294-1302. [DOI] [PubMed] [Google Scholar]
- 12.Goldmann, W., F. Houston, P. Stewart, M. Perucchini, J. Foster, and N. Hunter. 2006. Ovine prion protein variant A136R154L168Q171 increases resistance to experimental challenge with bovine spongiform encephalopathy agent. J. Gen. Virol. 87:3741-3745. [DOI] [PubMed] [Google Scholar]
- 13.Goldmann, W., N. Hunter, J. D. Foster, J. M. Salbaum, K. Beyreuther, and J. Hope. 1990. Two alleles of a neural protein gene linked to scrapie in sheep. Proc. Natl. Acad. Sci. USA 87:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heaton, M. P., K. A. Leymaster, B. A. Freking, D. A. Hawk, T. P. Smith, J. W. Keele, W. M. Snelling, J. M. Fox, C. G. Chitko-McKown, and W. W. Laegreid. 2003. Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle, and deer. Mamm. Genome 14:765-777. [DOI] [PubMed] [Google Scholar]
- 15.Houston, F., W. Goldmann, A. Chong, M. Jeffrey, L. Gonzalez, J. Foster, D. Parnham, and N. Hunter. 2003. Prion diseases: BSE in sheep bred for resistance to infection. Nature 423:498. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, N. 2003. Scrapie and experimental BSE in sheep. Br. Med. Bull. 66:171-183. [DOI] [PubMed] [Google Scholar]
- 17.Laplanche, J. L., J. Chatelain, D. Westaway, S. Thomas, M. Dussaucy, J. Brugere-Picoux, and J. M. Launay. 1993. PrP polymorphisms associated with natural scrapie discovered by denaturing gradient gel electrophoresis. Genomics 15:30-37. [DOI] [PubMed] [Google Scholar]
- 18.Moum, T., I. Olsaker, P. Hopp, T. Moldal, M. Valheim, T. Moum, and S. L. Benestad. 2005. Polymorphisms at codons 141 and 154 in the ovine prion protein gene are associated with scrapie Nor98 cases. J. Gen. Virol. 86:231-235. [DOI] [PubMed] [Google Scholar]
- 19.Piening, N., R. Nonno, M. Di Bari, S. Walter, O. Windl, U. Agrimi, H. A. Kretzschmar, and U. Bertsch. 2006. Conversion efficiency of bank vole prion protein in vitro is determined by residues 155 and 170, but does not correlate with the high susceptibility of bank voles to sheep scrapie in vivo. J. Biol. Chem. 281:9373-9384. [DOI] [PubMed] [Google Scholar]
- 20.Schutz, E., M. Scharfenstein, and B. Brenig. 2006. Genotyping of ovine prion protein gene (PRNP) variants by PCR with melting curve analysis. Clin. Chem. 52:1426-1429. [DOI] [PubMed] [Google Scholar]
- 21.Thorgeirsdottir, S., S. Sigurdarson, H. M. Thorisson, G. Georgsson, and A. Palsdottir. 1999. PrP gene polymorphism and natural scrapie in Icelandic sheep. J. Gen. Virol. 80:2527-2534. [DOI] [PubMed] [Google Scholar]
- 22.Vaccari, G., M. Conte, L. Morelli, G. Di Guardo, R. Petraroli, and U. Agrimi. 2004. Primer extension assay for prion protein genotype determination in sheep. Mol. Cell. Probes 18:33-37. [DOI] [PubMed] [Google Scholar]
- 23.Vaccari, G., M. A. Di Bari, L. Morelli, R. Nonno, B. Chiappini, G. Antonucci, S. Marcon, E. Esposito, P. Fazzi, N. Palazzini, P. Troiano, A. Petrella, G. Di Guardo, and U. Agrimi. 2006. Identification of an allelic variant of the goat PrP gene associated with resistance to scrapie. J. Gen. Virol. 87:1395-1402. [DOI] [PubMed] [Google Scholar]
- 24.Vaccari, G., R. Petraroli, U. Agrimi, C. Eleni, M. G. Perfetti, M. A. Di Bari, L. Morelli, C. Ligios, L. Busani, R. Nonno, and G. Di Guardo. 2001. PrP genotype in Sarda breed sheep and its relevance to scrapie. Arch. Virol. 146:2029-2037. [DOI] [PubMed] [Google Scholar]
- 25.Zhou, H., J. G. Hickford, and Q. Fang. 2005. Determination of alleles of the ovine PRNP gene using PCR-single-strand conformational polymorphism analysis. J. Anim. Sci. 83:745-749. [DOI] [PubMed] [Google Scholar]